Abstract
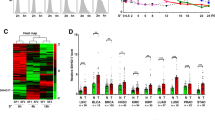
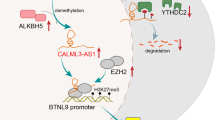
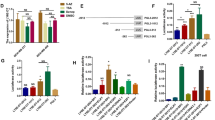
C-Myc overexpression contributes to multiple hallmarks of human cancer but directly targeting c-Myc is challenging. Identification of key factors involved in c-Myc dysregulation is of great significance to develop potential indirect targets for c-Myc. Herein, a collection of long non-coding RNAs (lncRNAs) interacted with c-Myc is detected in pancreatic ductal adenocarcinoma (PDAC) cells. Among them, lncRNA BCAN-AS1 is identified as the one with highest c-Myc binding enrichment. BCAN-AS1 was abnormally elevated in PDAC tumors and high BCAN-AS1 level was significantly associated with poor prognosis. Mechanistically, Smad nuclear-interacting protein 1 (SNIP1) was characterized as a new N6-methyladenosine (m6A) mediator binding to BCAN-AS1 via recognizing its m6A modification. m6A-modified BCAN-AS1 acts as a scaffold to facilitate the formation of a ternary complex together with c-Myc and SNIP1, thereby blocking S phase kinase-associated protein 2 (SKP2)-mediated c-Myc ubiquitination and degradation. Biologically, BCAN-AS1 promotes malignant phenotypes of PDAC in vitro and in vivo. Treatment of metastasis xenograft and patient-derived xenograft mouse models with in vivo-optimized antisense oligonucleotide of BCAN-AS1 effectively represses tumor growth and metastasis. These findings shed light on the pro-tumorigenic role of BCAN-AS1 and provide an innovant insight into c-Myc-interacted lncRNA in PDAC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
C-Myc RIP-seq, RNA-seq and SNIP1 CLIP-seq raw data generated in this study are publicly available in the Gene Expression Omnibus at GSE181777. miCLIP sequencing data have been deposited in the NCBI Short Read Archive with the BioProject ID-PRJNA693621. All custom code used to generate the data in this study is available upon reasonable request.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132.
Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–1039.
Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334.
van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309.
Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231.
Dhanasekaran R, Hansen AS, Park J, Lemaitre L, Lai I, Adeniji N, et al. MYC overexpression drives immune evasion in hepatocellular carcinoma that is reversible through restoration of proinflammatory macrophages. Cancer Res. 2023;83:626–640.
Sodir NM, Pellegrinet L, Kortlever RM, Campos T, Kwon YW, Kim S, et al. Reversible Myc hypomorphism identifies a key Myc-dependency in early cancer evolution. Nat Commun. 2022;13:6782.
Soucek L, Lawlor ER, Soto D, Shchors K, Swigart LB, Evan GI. Mast cells are required for angiogenesis and macroscopic expansion of Myc-induced pancreatic islet tumors. Nat Med. 2007;13:1211–1218.
Kortlever RM, Sodir NM, Wilson CH, Burkhart DL, Pellegrinet L, Brown Swigart L, et al. Myc cooperates with ras by programming inflammation and immune suppression. Cell. 2017;171:1301–1315.e1314.
Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35.
Hessmann E, Schneider G, Ellenrieder V, Siveke JT. MYC in pancreatic cancer: novel mechanistic insights and their translation into therapeutic strategies. Oncogene. 2016;35:1609–1618.
Skoudy A, Hernandez-Munoz I, Navarro P. Pancreatic ductal adenocarcinoma and transcription factors: role of c-Myc. J Gastrointest Cancer. 2011;42:76–84.
Sodir NM, Kortlever RM, Barthet VJA, Campos T, Pellegrinet L, Kupczak S, et al. MYC instructs and maintains pancreatic adenocarcinoma phenotype. Cancer Discov. 2020;10:588–607.
Maddipati R, Norgard RJ, Baslan T, Rathi KS, Zhang A, Saeid A, et al. MYC levels regulate metastatic heterogeneity in pancreatic adenocarcinoma. Cancer Discov. 2022;12:542–561.
Shih DJH, Nayyar N, Bihun I, Dagogo-Jack I, Gill CM, Aquilanti E, et al. Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat Genet. 2020;52:371–377.
Klotz R, Thomas A, Teng T, Han SM, Iriondo O, Li L, et al. Circulating tumor cells exhibit metastatic tropism and reveal brain metastasis drivers. Cancer Discov. 2020;10:86–103.
Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18:5546–5553.
Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104.
Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683.
Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5.
Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD, Hansen AS, Gouw AM, Felsher DW. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol. 2022;19:23–36.
Duffy MJ, Crown J. Drugging “undruggable” genes for cancer treatment: are we making progress? Int J Cancer. 2021;148:8–17.
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914.
Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62.
Huang H, Weng H, Chen J. m6A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–288.
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463.
Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261.
Iaccarino I. lncRNAs and MYC: an intricate relationship. Int J Mol Sci. 2017;18:1497.
Swier L, Dzikiewicz-Krawczyk A, Winkle M, van den Berg A, Kluiver J. Intricate crosstalk between MYC and non-coding RNAs regulates hallmarks of cancer. Mol Oncol. 2019;13:26–45.
Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86.
Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33:706–720.e709.
Hung CL, Wang LY, Yu YL, Chen HW, Srivastava S, Petrovics G, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci USA. 2014;111:18697–18702.
Luo L, Tang H, Ling L, Li N, Jia X, Zhang Z, et al. LINC01638 lncRNA activates MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation in triple-negative breast cancer. Oncogene. 2018;37:6166–6179.
Zhang Y, Gao L, Ma S, Ma J, Wang Y, Li S, et al. MALAT1-KTN1-EGFR regulatory axis promotes the development of cutaneous squamous cell carcinoma. Cell Death Differ. 2019;26:2061–2073.
Qu S, Jiao Z, Lu G, Yao B, Wang T, Rong W, et al. PD-L1 lncRNA splice isoform promotes lung adenocarcinoma progression via enhancing c-Myc activity. Genome Biol. 2021;22:104.
Shen S, Yao T, Xu Y, Zhang D, Fan S, Ma J. CircECE1 activates energy metabolism in osteosarcoma by stabilizing c-Myc. Mol Cancer. 2020;19:151.
Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–1620.
Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–446.
Sobin LH, Compton CC. TNM seventh edition: what’s new, what’s changed: communication from the International Union Against Cancer and the American Joint Committee on Cancer. Cancer. 2010;116:5336–5339.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550.
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87.
Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10:1858.
Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc. 2014;9:263–293.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760.
Shah A, Qian Y, Weyn-Vanhentenryck SM, Zhang C. CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics. 2017;33:566–567.
Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–772.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359.
Uren PJ, Bahrami-Samani E, Burns SC, Qiao M, Karginov FV, Hodges E, et al. Site identification in high-throughput RNA-protein interaction data. Bioinformatics. 2012;28:3013–3020.
Robinson J, Thorvaldsdóttir H, Winckler W, Guttman M, Lander E, Getz G, et al. Integrated genomics viewer. Nat Biotechnol. 2011;29:24–26.
Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74.
Deng J, Zhang J, Ye Y, Liu K, Zeng L, Huang J, et al. N6-methyladenosine-mediated upregulation of WTAPP1 promotes WTAP translation and wnt signaling to facilitate pancreatic cancer progression. Cancer Res. 2021;81:5268–5283.
Muppirala UK, Honavar VG, Dobbs D. Predicting RNA-protein interactions using only sequence information. BMC Bioinforma. 2011;12:489.
Farrell AS, Sears RC. MYC degradation. Cold Spring Harb Perspect Med. 2014;4:a014365.
Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11:1177–1188.
von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–1200.
Fujii M, Lyakh LA, Bracken CP, Fukuoka J, Hayakawa M, Tsukiyama T, et al. SNIP1 is a candidate modifier of the transcriptional activity of c-Myc on E box-dependent target genes. Mol Cell. 2006;24:771–783.
Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–345.
Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–1205.
Li J, Chen Z, Chen F, Xie G, Ling Y, Peng Y, et al. Targeted mRNA demethylation using an engineered dCas13b-ALKBH5 fusion protein. Nucleic Acids Res. 2020;48:5684–5694.
Li Z, Peng Y, Li J, Chen Z, Chen F, Tu J, et al. N6-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun. 2020;11:2578.
Shao B, Fu X, Li X, Li Y, Gan N. RP11-284F21.9 promotes oral squamous cell carcinoma development via the miR-383-5p/MAL2 axis. J Oral Pathol Med. 2020;49:21–29.
Li D, Wang L, Feng J, Shen YW, Liu LN, Wang Y. RP11‑284F21.9 promotes lung carcinoma proliferation and invasion via the regulation of miR‑627‑3p/CCAR1. Oncol Rep. 2020;44:1638–1648.
Han HF, Chen Q, Zhao WW. Long non-coding RNA RP11-284F21.9 functions as a ceRNA regulating PPWD1 by competitively binding to miR-769-3p in cervical carcinoma. Biosci. Rep. 2020;40:BSR20200784.
Roche KC, Wiechens N, Owen-Hughes T, Perkins ND. The FHA domain protein SNIP1 is a regulator of the cell cycle and cyclin D1 expression. Oncogene. 2004;23:8185–8195.
Kim RH, Flanders KC, Birkey Reffey S, Anderson LA, Duckett CS, Perkins ND, et al. SNIP1 inhibits NF-kappa B signaling by competing for its binding to the C/H1 domain of CBP/p300 transcriptional co-activators. J Biol Chem. 2001;276:46297–46304.
Chen LL, Lin HP, Zhou WJ, He CX, Zhang ZY, Cheng ZL, et al. SNIP1 recruits TET2 to regulate c-MYC target genes and cellular DNA damage response. Cell Rep. 2018;25:1485–1500.e1484.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641.
Zhong Y, Yang L, Xiong F, He Y, Tang Y, Shi L, et al. Long non-coding RNA AFAP1-AS1 accelerates lung cancer cells migration and invasion by interacting with SNIP1 to upregulate c-Myc. Signal Transduct Target Ther. 2021;6:240.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Zhou KR, Liu S, Sun WJ, Zheng LL, Zhou H, Yang JH, et al. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017;45:D43–D50.
Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200.
Lee J, Wu Y, Harada BT, Li Y, Zhao J, He C et al. N6-methyladenosine modification of lncRNA Pvt1 governs epidermal stemness. EMBO J. 2021;40:e106276.
Acknowledgements
This study was supported by the National Key R&D Program of China (2021YFA1302100 to J. Zheng), National Natural Science Foundation of China (82325037, 82072617 to J. Zheng and 82003162 to J. Zhang), Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2017ZT07S096 to D.L.), China Postdoctoral Science Foundation (2023M734042 to J.S.) and Sun Yat-sen University Intramural Funds (to D.L. and to J. Zheng).
Author information
Authors and Affiliations
Contributions
J. Zheng and J. Zhang conceptualized and supervised this study. G.W., J.S. and L. Zeng performed most experiments. S.D., Y.Y. and R.L. conducted statistical and bioinformatics analyses. J. Zhang and X.H. performed animal experiments. Q.Z., Y.Z., J.D., S.Z. and R.C. contributed to collection of clinic samples. M.L. contributed to histopathological analyses. R.B. and L. Zhuang provided technique supports. G.W. J.S., J. Zheng and D.L. prepared manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was approved by the Institutional Review Board of the Sun Yat-sen Memorial Hospital and Chinese Academy of Medical Sciences Cancer Hospital. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Sun Yat-sen University Cancer Center and the animals were handled in accordance with institutional guidelines.
Informed consent
Informed consent was obtained from each patient.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, G., Su, J., Zeng, L. et al. LncRNA BCAN-AS1 stabilizes c-Myc via N6-methyladenosine-mediated binding with SNIP1 to promote pancreatic cancer. Cell Death Differ 30, 2213–2230 (2023). https://doi.org/10.1038/s41418-023-01225-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01225-x