Isolation, Identification and Investigation of Fermentative Bacteria from Sea Bass (Dicentrarchus labrax): Evaluation of Antifungal Activity of Fermented Fish Meat and By-Products Broths
Abstract
:1. Introduction
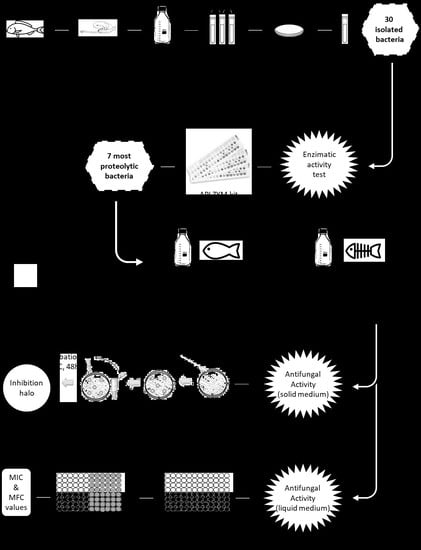
2. Materials and Methods
2.1. Materials
2.1.1. Samples
2.1.2. Microorganisms
2.1.3. Consumables
2.1.4. Fish By-Products Broths
2.2. Methods
2.2.1. Isolation and Identification of Bacteria by Morphology, Catalase Test and PCR
2.2.2. Enzymatic Activity
2.2.3. Fermentation of Isolated Bacteria in Meat and By-Products Broths
2.2.4. Antifungal Activity
2.3. Statistical Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2018: Meeting the Sustainable Development Goals. Available online: http://www.fao.org/3/i9540en/i9540en.pdf (accessed on 30 March 2020).
- Olsen, R.L.; Toppe, J.; Karunasagar, I. Challenges and realistic opportunities in the use of by-products from processing of fish and shellfish. Trends Food Sci. Technol. 2014, 36, 144–151. [Google Scholar] [CrossRef]
- Martínez-Alvarez, O.; Chamorro, S.; Brenes, A. Protein hydrolysates from animal processing by-products as a source of bioactive molecules with interest in animal feeding: A review. Food Res. Int. 2015, 73, 204–212. [Google Scholar]
- Al Khawli, F.; Pateiro, M.; Domínguez, R.; Lorenzo, J.M.; Gullón, P.; Kousoulaki, K.; Ferrer, E.; Berrada, H.; Barba, F.J. Innovative green technologies of intensification for valorization of seafood and their by-products. Mar. Drugs 2019, 17, 689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez, B.; Munekata, P.E.S.; Gavahian, M.; Barba, F.J.; Martí-Quijal, F.J.; Bolumar, T.; Campagnol, P.C.B.; Tomasevic, I.; Lorenzo, J.M. Application of pulsed electric fields in meat and fish processing industries: An overview. Food Res. Int. 2019, 123, 95–105. [Google Scholar]
- Bruno, S.F.; Ekorong, F.J.A.A.; Karkal, S.S.; Cathrine, M.S.B.; Kudre, T.G. Green and innovative techniques for recovery of valuable compounds from seafood by-products and discards: A review. Trends Food Sci. Technol. 2019, 85, 10–22. [Google Scholar] [CrossRef]
- Marti-Quijal, F.J.; Remize, F.; Meca, G.; Ferrer, E.; Ruiz, M.-J.; Barba, F.J. Fermentation in fish and by-products processing: An overview of current research and future prospects. Curr. Opin. Food Sci. 2020, 31, 9–16. [Google Scholar] [CrossRef]
- Zang, J.; Xu, Y.; Xia, W.; Regenstein, J.M. Quality, functionality, and microbiology of fermented fish: A review. Crit. Rev. Food Sci. Nutr. 2019, 60, 1228–1242. [Google Scholar] [CrossRef]
- Rai, A.K.; Jini, R.; Swapna, H.C.; Sachindra, N.M.; Bhaskar, N.; Baskaran, V. Application of native lactic acid bacteria (LAB) for fermentative recovery of lipids and proteins from fish processing wastes: Bioactivities of fermentation products. J. Aquat. Food Prod. Technol. 2011, 20, 32–44. [Google Scholar] [CrossRef]
- Rai, A.K.; Bhaskar, N.; Halami, P.M.; Indirani, K.; Suresh, P.V.; Mahendrakar, N.S. Characterization and application of a native lactic acid bacterium isolated from tannery fleshings for fermentative bioconversion of tannery fleshings. Appl. Microbiol. Biotechnol. 2009, 83, 757–766. [Google Scholar] [CrossRef]
- Rai, A.K.; Swapna, H.C.; Bhaskar, N.; Halami, P.M.; Sachindra, N.M. Effect of fermentation ensilaging on recovery of oil from fresh water fish viscera. Enzyme Microb. Technol. 2010, 46, 9–13. [Google Scholar] [CrossRef]
- Murthy, P.S.; Rai, A.K.; Bhaskar, N. Fermentative recovery of lipids and proteins from freshwater fish head waste with reference to antimicrobial and antioxidant properties of protein hydrolysate. J. Food Sci. Technol. 2014, 51, 1884–1892. [Google Scholar]
- Harkin, C.; Bruck, W.M.; Lynch, C. Isolation & identification of bacteria for the treatment of brown crab (Cancer pagurus) waste to produce chitinous material. J. Appl. Microbiol. 2015, 118, 954–965. [Google Scholar] [PubMed]
- Pacheco, N.; Garnica-González, M.; Ramírez-Hernández, J.Y.; Flores-Albino, B.; Gimeno, M.; Bárzana, E.; Shirai, K. Effect of temperature on chitin and astaxanthin recoveries from shrimp waste using lactic acid bacteria. Bioresour. Technol. 2009, 100, 2849–2854. [Google Scholar] [CrossRef] [PubMed]
- Martí-Quijal, F.J.; Tornos, A.; Príncep, A.; Luz, C.; Meca, G.; Tedeschi, P.; Ruiz, M.-J.; Barba, F.J. Impact of fermentation on the recovery of antioxidant bioactive compounds from sea bass byproducts. Antioxidants 2020, 9, 239. [Google Scholar]
- Crowley, S.; Mahony, J.; Van Sinderen, D. Current perspectives on antifungal lactic acid bacteria as natural bio-preservatives. Trends Food Sci. Technol. 2013, 33, 93–109. [Google Scholar] [CrossRef]
- Gerez, C.L.; Fornaguera, M.J.; Obregozo, M.D.; de Valdez, G.F.; Torino, M.I. Antifungal starter culture for packed bread: Influence of two storage conditions. Rev. Argent. Microbiol. 2015, 47, 118–124. [Google Scholar] [CrossRef] [Green Version]
- Arora, G.; Lee, B.H.; Lamoureux, M. Characterization of enzyme profiles of Lactobacillus casei species by a rapid API ZYM system. J. Dairy Sci. 1990, 73, 264–273. [Google Scholar] [CrossRef]
- Altınelataman, C.; Koroleva, O.; Fedorova, T.; Torkova, A.; Lisitskaya, K.; Tsentalovich, M.; Kononikhin, A.; Popov, I.; Vasina, D.; Kovalyov, L.; et al. An in vitro and in silico study on the antioxidant and cell culture-based study on the chemoprotective activities of fish muscle protein hydrolysates obtained from European seabass and gilthead seabream. Food Chem. 2019, 271, 724–732. [Google Scholar]
- Varsha, K.K.; Priya, S.; Devendra, L.; Nampoothiri, K.M. Control of spoilage fungi by protective lactic acid bacteria displaying probiotic properties. Appl. Biochem. Biotechnol. 2014, 172, 3402–3413. [Google Scholar] [CrossRef]
- Fothergill, A.W. Antifungal Susceptibility Testing: Clinical Laboratory and Standards Institute (CLSI) Methods; Humana Press: Totowa, NJ, USA, 2012; Volume 9781597451345, pp. 65–74. [Google Scholar]
- Axelsson, L. Lactic acid bacteria: Classification and physiology. In Lactic Acid Bacteria Microbiological and Functional Aspects, Third Edition: Revised and Expanded; CRC Press: Boca Raton, FL, USA, 2004; pp. 1–66. [Google Scholar]
- Gerbaldo, G.A.; Barberis, C.; Pascual, L.; Dalcero, A.; Barberis, L. Antifungal activity of two Lactobacillus strains with potential probiotic properties. FEMS Microbiol. Lett. 2012, 332, 27–33. [Google Scholar] [CrossRef]
- Bajpai, V.K.; Han, J.-H.; Nam, G.-J.; Majumder, R.; Park, C.; Lim, J.; Paek, W.K.; Rather, I.A.; Park, Y.-H. Characterization and pharmacological potential of Lactobacillus sakei 1I1 isolated from fresh water fish Zacco koreanus. Daru 2016, 24, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leyva Salas, M.; Mounier, J.; Valence, F.; Coton, M.; Thierry, A.; Coton, E. Antifungal microbial agents for food biopreservation—A review. Microorganisms 2017, 5, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Liu, J.; Li, J.; Chen, C.; Zhang, H.; Wang, H.-K.; Lu, F.-P. Characteristics and application in food preservatives of Lactobacillus plantarum TK9 isolated from naturally fermented congee. Int. J. Food Eng. 2016, 12, 377–384. [Google Scholar] [CrossRef]
- Cray, J.A.; Bell, A.N.W.; Bhaganna, P.; Mswaka, A.Y.; Timson, D.J.; Hallsworth, J.E. The biology of habitat dominance; can microbes behave as weeds? Microb. Biotechnol. 2013, 6, 453–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, E.H.; Yang, E.J.; Woo, E.R.; Chang, H.C. Purification and characterization of antifungal compounds from Lactobacillus plantarum HD1 isolated from kimchi. Food Microbiol. 2014, 41, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Ringø, E.; Bendiksen, H.R.; Wesmajervi, M.S.; Olsen, R.E.; Jansen, P.A.; Mikkelsen, H. Lactic acid bacteria associated with the digestive tract of Atlantic salmon (Salmo salar L.). J. Appl. Microbiol. 2000, 89, 317–322. [Google Scholar]
- Ringø, E.; Bendiksen, H.R.; Gausen, S.J.; Sundsfjord, A.; Olsen, R.E. The effect of dietary fatty acids on lactic acid bacteria associated with the epithelial mucosa and from faecalia of Arctic charr, Salvelinus alpinus (L.). J. Appl. Microbiol. 1998, 85, 855–864. [Google Scholar]
- Yimin, C.; Suyanandana, P.; Saman, P.; Benno, Y. Classification and characterization of lactic acid bacteria isolated from the intestines of common carp and freshwater prawns. J. Gen. Appl. Microbiol. 1999, 45, 177–184. [Google Scholar]
- Strom, E.; Olafsen, J. Microbiology in Poecilotherm; Lésel, R., Ed.; Elsevier: Amsterdam, The Netherlands, 1990; pp. 181–185. [Google Scholar]
- Hagi, T.; Tanaka, D.; Iwamura, Y.; Hoshino, T. Diversity and seasonal changes in lactic acid bacteria in the intestinal tract of cultured freshwater fish. Aquaculture 2004, 234, 335–346. [Google Scholar] [CrossRef]
- Nair, P.; Surendran, P. Biochemical characterization of lactic acid bacteria isolated from fish prawn. J. Cult. Collect. 2005, 4, 48–52. [Google Scholar]
- Sahnouni, F.; Ringø, E.; Maizi, A.; Belmaghnia, S.; Matallah-Boutiba, A.; Chemlal, D.; Boutiba, Z. Biochemical and antibacterial potential of autochthonous carnobacterium and lactobacillus species isolated from gastrointestinal tract of coastal fish. J. Anim. Plant Sci. 2016, 26, 1146–1155. [Google Scholar]
- Gatesoupe, F.-J. Updating the importance of lactic acid bacteria in fish farming: Natural occurrence and probiotic treatments. J. Mol. Microbiol. Biotechnol. 2007, 14, 107–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, S.; Carmen Castro, M.; Berdasco, M.; de la Banda, I.G.; Moreno-Ventas, X.; de Rojas, A.H. Isolation and partial characterization of lactic acid bacteria from the gut microbiota of marine fishes for potential application as probiotics in aquaculture. Probiotics Antimicrob. Proteins 2019, 11, 569–579. [Google Scholar] [CrossRef] [PubMed]




| Enzymes | Stomach Bacteria | Small Intestine Bacteria | Colon Bacteria | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | S4 | S5 | S6 | S7 | I1 | I2 | I3 | I4 | I5 | I6 | I7 | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 | C11 | C12 | C13 | C14 | C15 | C16 | |
| Alkaline Phosphatase | 10 | ≥40 | ≥40 | ≥40 | ≥40 | 5 | 5 | 5 | 5 | 5 | 0 | 30 | 5 | 5 | 20 | 10 | 10 | 20 | 10 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 10 | 5 | 5 | 0 |
| Esterase (C4) | 5 | 20 | 20 | 20 | 5 | ≥40 | ≥40 | 20 | 10 | 5 | 0 | 20 | 10 | 5 | 0 | 20 | 30 | 20 | 20 | 20 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 5 | 5 |
| Esterase Lipase (C8) | 5 | 20 | 10 | 10 | 10 | 20 | 30 | 20 | 30 | 0 | 0 | 10 | 20 | 20 | 0 | 20 | 10 | 10 | 10 | 10 | 10 | 5 | 0 | 0 | 5 | 0 | 5 | 5 | 5 | 5 |
| Lipase (C14) | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Leucine Arylamidase | ≥40 | 30 | 30 | 30 | 20 | 30 | 20 | 0 | 5 | 5 | 0 | 30 | ≥40 | 10 | 20 | 30 | 30 | ≥40 | 30 | 30 | 20 | 0 | 5 | 0 | 0 | 0 | ≥40 | 0 | 10 | 5 |
| Valine Arylamidase | 30 | 5 | 0 | 0 | 0 | 5 | 0 | 5 | 5 | 5 | 0 | 0 | ≥40 | 10 | 0 | 5 | 0 | 20 | 5 | 0 | 5 | 0 | 5 | 0 | 0 | 0 | ≥40 | 10 | 10 | 5 |
| Cystine Arylamidase | 20 | 0 | 0 | 10 | 0 | 0 | 10 | 10 | 5 | 0 | 0 | 5 | 10 | 0 | 5 | 10 | 0 | 5 | 0 | 0 | 0 | 10 | 0 | 5 | 0 | 5 | 10 | 10 | 10 | 0 |
| Trypsin | 0 | 5 | 0 | 10 | 0 | 10 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 5 | 10 | 10 | 5 |
| α-chymotrypsin | 0 | 0 | 10 | 5 | 0 | 0 | 10 | 10 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 |
| Acid Phosphatase | 30 | 10 | 10 | 10 | 10 | 5 | 5 | 10 | 5 | 0 | 5 | 5 | ≥40 | 10 | 5 | 5 | 0 | 10 | 5 | 10 | 10 | 20 | 5 | 10 | 5 | 10 | ≥40 | 30 | 30 | 5 |
| Naphthol-as-bi-phosphohydrolase | 30 | 10 | 5 | 10 | 20 | 5 | 10 | 10 | 5 | 5 | 5 | 10 | ≥40 | 10 | 5 | 10 | 20 | 10 | 5 | 10 | 10 | 10 | 5 | 20 | 10 | 5 | 30 | 30 | 20 | 10 |
| α-Galactosidase | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | ≥40 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| β-Galactosidase | 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ≥40 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | ≥40 | 0 | 0 | 5 |
| β-Glucuronidase | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ≥40 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| α-Glucosidase | 20 | 10 | 10 | 10 | 10 | 5 | 0 | 0 | 0 | 5 | 0 | 0 | ≥40 | 0 | 0 | ≥40 | 0 | 5 | 10 | ≥40 | 30 | ≥40 | ≥40 | 5 | 0 | ≥40 | 10 | ≥40 | ≥40 | 5 |
| β-Glucosidase | ≥40 | 10 | 5 | 0 | 10 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | ≥40 | 0 | 0 | 30 | 0 | 0 | 20 | ≥40 | 30 | 0 | 0 | 30 | ≥40 | 0 | ≥40 | 0 | 0 | ≥40 |
| N-Acethyl-β-glucosaminidase | 0 | 0 | 5 | 0 | 0 | 0 | 5 | 0 | 5 | 0 | 0 | 5 | 0 | 10 | 5 | 10 | 20 | 0 | 0 | 0 | 0 | 10 | 5 | 5 | 0 | 0 | 0 | 5 | 0 | 5 |
| α-Mannosidase | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| α-Fucosidase | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 |
| Fungi | Control | S3 | S4 | S6 | S7 | I1 | C14 | C15 |
|---|---|---|---|---|---|---|---|---|
| Meat Broth | ||||||||
| Aspergillus parasiticus | - | +++ | - | - | - | - | - | - |
| Aspergillus flavus | - | + | - | - | - | - | - | - |
| Penicillium expansum | - | +++ | - | - | - | - | - | - |
| Penicillium verrucosum | - | - | - | - | - | - | + | - |
| Fusarium graminearum | - | +++ | - | - | - | - | + | - |
| Fusarium verticillioides | - | +++ | - | - | - | - | - | - |
| By-Products Broth | ||||||||
| Aspergillus parasiticus | - | +++ | ++ | - | - | - | - | - |
| Aspergillus flavus | - | + | + | - | - | - | - | - |
| Penicillium expansum | - | +++ | +++ | - | + | - | - | - |
| Penicillium verrucosum | - | + | + | - | - | - | - | - |
| Fusarium graminearum | - | + | + | - | - | - | + | - |
| Fusarium verticillioides | - | +++ | +++ | - | - | - | - | - |
| Fungi | C14 | S3 | S4 | C14 | ||||
|---|---|---|---|---|---|---|---|---|
| MIC | MFC | MIC | MFC | MIC | MFC | MIC | MFC | |
| Meat Broth | Waste Broth | |||||||
| Aspergillus parasiticus | nd | nd | 8 | nd | 16 | nd | nd | nd |
| Aspergillus flavus | nd | nd | 16 | nd | 32 | nd | nd | nd |
| Penicillum expansum | nd | nd | 1 | 16 | 8 | 16 | nd | nd |
| Penicillium verrucosum | 16 | 32 | 4 | 8 | 4 | 16 | nd | nd |
| Fusarium graminearum | 16 | nd | 2 | 16 | 1 | 31 | 16 | nd |
| Fusarium verticillioides | nd | nd | 1 | 8 | 4 | 16 | nd | nd |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martí-Quijal, F.J.; Príncep, A.; Tornos, A.; Luz, C.; Meca, G.; Tedeschi, P.; Ruiz, M.-J.; Barba, F.J.; Mañes, J. Isolation, Identification and Investigation of Fermentative Bacteria from Sea Bass (Dicentrarchus labrax): Evaluation of Antifungal Activity of Fermented Fish Meat and By-Products Broths. Foods 2020, 9, 576. https://doi.org/10.3390/foods9050576
Martí-Quijal FJ, Príncep A, Tornos A, Luz C, Meca G, Tedeschi P, Ruiz M-J, Barba FJ, Mañes J. Isolation, Identification and Investigation of Fermentative Bacteria from Sea Bass (Dicentrarchus labrax): Evaluation of Antifungal Activity of Fermented Fish Meat and By-Products Broths. Foods. 2020; 9(5):576. https://doi.org/10.3390/foods9050576
Chicago/Turabian StyleMartí-Quijal, Francisco J., Andrea Príncep, Adrián Tornos, Carlos Luz, Giuseppe Meca, Paola Tedeschi, María-José Ruiz, Francisco J. Barba, and Jordi Mañes. 2020. "Isolation, Identification and Investigation of Fermentative Bacteria from Sea Bass (Dicentrarchus labrax): Evaluation of Antifungal Activity of Fermented Fish Meat and By-Products Broths" Foods 9, no. 5: 576. https://doi.org/10.3390/foods9050576