Evaluation of Dimercaptosuccinic Acid-Coated Iron Nanoparticles Immunotargeted to Amyloid Beta as MRI Contrast Agents for the Diagnosis of Alzheimer’s Disease
Abstract
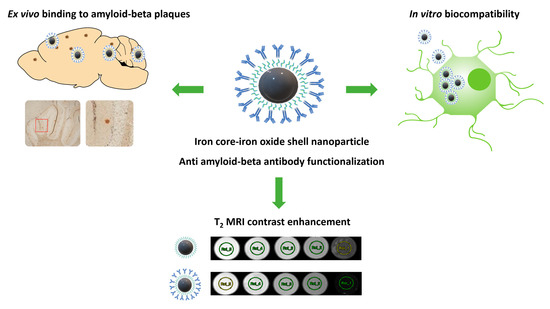
:1. Introduction
2. Materials and Methods
2.1. Synthesis of Iron Nanoparticles
2.2. Particle Size and Zeta Potential
2.3. Conjugation of Anti-Amyloid Antibodies to the Surface of Nanospheres
2.4. MRI of Nanoparticles
2.5. Cell Culture
2.6. Lactate Dehydrogenase Assay
2.7. Ames Bacterial Mutation Assay
2.8. Determination of Reactive Oxygen Species/Superoxide Production
2.9. Quantification of Internalized Nanoparticles Using Inductively Coupled Plasma-Mass-Spectrometry
2.10. TEM Analysis of Nanoparticle Internalization into Cells
2.11. Immunohistochemistry
2.12. U-251 Cell Proteomics
2.12.1. Cell Culture and Nanoparticle Treatment
2.12.2. Cell Lysis and Protein Digestion
2.12.3. Tandem Mass Spectrometry and Database Searching
2.12.4. Bioinformatics and Statistical Analyses
2.13. Animals
Immunohistochemical Characterization of NP-Ab
3. Results
3.1. Nanoparticle Characterization
3.2. Cell Viability
3.3. ROS/Superoxide Production
3.4. Mutagenicity of Iron Nanoparticles and NP-Ab in the Ames Test
3.5. Cellular Internalization of Iron Nanoparticles and NP-Ab
3.6. MRI of Iron Nanoparticles and NP-Ab
3.7. Proteomics
Gene Ontology Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Dementia. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed on 9 July 2023).
- Hillen, H. The Beta Amyloid Dysfunction (BAD) Hypothesis for Alzheimer’s Disease. Hypothesis and Theory. Front. Neurosci. 2019, 13, 1154. [Google Scholar] [CrossRef] [PubMed]
- Blennow, K.; Dubois, B.; Fagan, A.M.; Lewczuk, P.; de Leon, M.J.; Hampel, H. Clinical utility of cerebrospinal fluid biomarkers in the diagnosis of early Alzheimer’s disease. Alzheimers Dement. 2015, 11, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Vogelgsang, J.; Wedekind, D.; Bouter, C.; Klafki, H.W.; Wiltfang, J. Reproducibility of Alzheimer’s Disease Cerebrospinal Fluid-Biomarker Measurements under Clinical Routine Conditions. J. Alzheimers Dis. 2018, 62, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Moses, W.W. Fundamental Limits of Spatial Resolution in PET. Nucl. Instrum. Methods Phys. Res. Sect. A 2011, 648 (Suppl. S1), S236–S240. [Google Scholar] [CrossRef] [PubMed]
- Petiet, A.; Santin, M.; Bertrand, A.; Wiggins, C.J.; Petit, F.; Houitte, D.; Hantraye, P.; Benavides, J.; Debeir, T.; Rooney, T.; et al. Gadolinium-staining reveals amyloid plaques in the brain of Alzheimer’s transgenic mice. Neurobiol. Aging 2012, 33, 1533–1544. [Google Scholar] [CrossRef]
- Dudeffant, C.; Vandesquille, M.; Herbert, K.; Garin, C.M.; Alves, S.; Blanchard, V.; Comoy, E.E.; Petit, F.; Dhenain, M. Contrast-enhanced MR microscopy of amyloid plaques in five mouse models of amyloidosis and in human Alzheimer’s disease brains. Sci. Rep. 2017, 7, 4955. [Google Scholar] [CrossRef]
- Badachhape, A.A.; Working, P.K.; Srivastava, M.; Bhandari, P.; Stupin, I.V.; Devkota, L.; Tanifum, E.A.; Annapragada, A.V.; Ghaghada, K.B. Pre-clinical dose-ranging efficacy, pharmacokinetics, tissue biodistribution, and toxicity of a targeted contrast agent for MRI of amyloid deposition in Alzheimer’s disease. Sci. Rep. 2020, 10, 16185. [Google Scholar] [CrossRef]
- Wang, X.; Chan, H.N.; Desbois, N.; Gros, C.P.; Bolze, F.; Li, Y.; Li, H.W.; Wong, M.S. Multimodal Theranostic Cyanine-Conjugated Gadolinium(III) Complex for In Vivo Imaging of Amyloid-β in an Alzheimer’s Disease Mouse Model. ACS Appl. Mater. Interfaces 2021, 13, 18525–18532. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Chan, H.-N.; Liu, G.; Wang, Z.; Li, H.-W.; Wong, M.S. Amyloid-β Oligomer-Targeted Gadolinium-Based NIR/MR Dual-Modal Theranostic Nanoprobe for Alzheimer’s Disease. Adv. Funct. Mater. 2020, 30, 1909529. [Google Scholar] [CrossRef]
- Cowper, S.E.; Robin, H.S.; Steinberg, S.M.; Su, L.D.; Gupta, S.; LeBoit, P.E. Scleromyxoedema-like cutaneous diseases in renal-dialysis patients. Lancet 2000, 356, 1000–1001. [Google Scholar] [CrossRef]
- Grobner, T. Gadolinium--a specific trigger for the development of nephrogenic fibrosing dermopathy and nephrogenic systemic fibrosis? Nephrol. Dial. Transplant. 2006, 21, 1104–1108. [Google Scholar] [CrossRef] [PubMed]
- Marckmann, P.; Skov, L.; Rossen, K.; Dupont, A.; Damholt, M.B.; Heaf, J.G.; Thomsen, H.S. Nephrogenic Systemic Fibrosis: Suspected Causative Role of Gadodiamide Used for Contrast-Enhanced Magnetic Resonance Imaging. Am. J. Nephrol. 2006, 17, 2359. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Ishii, K.; Kawaguchi, H.; Kitajima, K.; Takenaka, D. High signal intensity in the dentate nucleus and globus pallidus on unenhanced T1-weighted MR images: Relationship with increasing cumulative dose of a gadolinium-based contrast material. Radiology 2014, 270, 834–841. [Google Scholar] [CrossRef] [PubMed]
- Murata, N.; Gonzalez-Cuyar, L.F.; Murata, K.; Fligner, C.; Dills, R.; Hippe, D.; Maravilla, K.R. Macrocyclic and Other Non-Group 1 Gadolinium Contrast Agents Deposit Low Levels of Gadolinium in Brain and Bone Tissue: Preliminary Results From 9 Patients With Normal Renal Function. Investig. Radiol. 2016, 51, 447–453. [Google Scholar] [CrossRef]
- Yeo, S.K.; Shepelytskyi, Y.; Grynko, V.; Albert, M.S. Molecular Imaging of Fluorinated Probes for Tau Protein and Amyloid-β Detection. Molecules 2020, 25, 3413. [Google Scholar] [CrossRef]
- Yanagisawa, D.; Ibrahim, N.F.; Taguchi, H.; Morikawa, S.; Tomiyama, T.; Tooyama, I. Fluorine-19 Magnetic Resonance Imaging for Detection of Amyloid β Oligomers Using a Keto Form of Curcumin Derivative in a Mouse Model of Alzheimer’s Disease. Molecules 2021, 26, 1362. [Google Scholar] [CrossRef]
- Yousaf, M.; Ahmad, M.; Bhatti, I.A.; Nasir, A.; Hasan, M.; Jian, X.; Kalantar-Zadeh, K.; Mahmood, N. In Vivo and In Vitro Monitoring of Amyloid Aggregation via BSA@FGQDs Multimodal Probe. ACS Sens. 2019, 4, 200–210. [Google Scholar] [CrossRef]
- Ulanova, M.; Poljak, A.; Wen, W.; Bongers, A.; Gloag, L.; Gooding, J.; Tilley, R.; Sachdev, P.; Braidy, N. Nanoparticles as contrast agents for the diagnosis of Alzheimer’s disease: A systematic review. Nanomedicine 2020, 15, 725–743. [Google Scholar] [CrossRef]
- Zhou, J.; Fa, H.; Yin, W.; Zhang, J.; Hou, C.; Huo, D.; Zhang, D.; Zhang, H. Synthesis of superparamagnetic iron oxide nanoparticles coated with a DDNP-carboxyl derivative for in vitro magnetic resonance imaging of Alzheimer’s disease. Mater. Sci. Eng. C Mater. Biol. Appl. 2014, 37, 348–355. [Google Scholar] [CrossRef]
- Zhang, D.; Fa, H.B.; Zhou, J.T.; Li, S.; Diao, X.W.; Yin, W. The detection of β-amyloid plaques in an Alzheimer’s disease rat model with DDNP-SPIO. Clin. Radiol. 2015, 70, 74–80. [Google Scholar] [CrossRef]
- Tsolakis, A.C.; Halevas, E.; Vouroutzis, N.; Koliakos, G.G.; Salifoglou, A.; Litsardakis, G. Magnetic Fluorescent Nanoparticles Binding to Amyloid-Beta Peptide: Silica-Coated, Thioflavin-T Functionalized Iron Oxide. IEEE Trans. Magn. 2017, 53, 5300804. [Google Scholar] [CrossRef]
- Wadghiri, Y.Z.; Sigurdsson, E.M.; Sadowski, M.; Elliott, J.I.; Li, Y.; Scholtzova, H.; Tang, C.Y.; Aguinaldo, G.; Pappolla, M.; Duff, K.; et al. Detection of Alzheimer’s amyloid in transgenic mice using magnetic resonance microimaging. Magn. Reson. Med. 2003, 50, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wadghiri, Y.Z.; Hoang, D.M.; Tsui, W.; Sun, Y.; Chung, E.; Li, Y.; Wang, A.; de Leon, M.; Wisniewski, T. Detection of amyloid plaques targeted by USPIO-Aβ1-42 in Alzheimer’s disease transgenic mice using magnetic resonance microimaging. Neuroimage 2011, 55, 1600–1609. [Google Scholar] [CrossRef] [PubMed]
- Wadghiri, Y.Z.; Li, J.; Wang, J.; Hoang, D.M.; Sun, Y.; Xu, H.; Tsui, W.; Li, Y.; Boutajangout, A.; Wang, A.; et al. Detection of amyloid plaques targeted by bifunctional USPIO in Alzheimer’s disease transgenic mice using magnetic resonance microimaging. PLoS ONE 2013, 8, e57097. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Qu, Y.; Min, Z.; Wu, J.; Zhang, S.; Xue, Z. Amyloid Plaque Imaging with a Targeted MRI Contrast Agent in a Transgenic Mouse Model of Alzheimer’s Disease. Int. J. Nanomed. 2022, 17, 927–936. [Google Scholar] [CrossRef]
- Viola, K.L.; Sbarboro, J.; Sureka, R.; De, M.; Bicca, M.A.; Wang, J.; Vasavada, S.; Satpathy, S.; Wu, S.; Joshi, H.; et al. Towards non-invasive diagnostic imaging of early-stage Alzheimer’s disease. Nat. Nanotechnol. 2015, 10, 91–98. [Google Scholar] [CrossRef]
- Yin, Z.; Yul, T.; Xu, Y. Preparation of Amyloid Immuno-Nanoparticles as Potential MRI Contrast Agents for Alzheimer’s Disease Diagnosis. J. Nanosci. Nanotechnol. 2015, 15, 6429–6434. [Google Scholar] [CrossRef]
- Solberg, N.O.; Chamberlin, R.; Vigil, J.R.; Deck, L.M.; Heidrich, J.E.; Brown, D.C.; Brady, C.I.; Vander Jagt, T.A.; Garwood, M.; Bisoffi, M.; et al. Optical and SPION-enhanced MR imaging shows that trans-stilbene inhibitors of NF-κB concomitantly lower Alzheimer’s disease plaque formation and microglial activation in AβPP/PS-1 transgenic mouse brain. J. Alzheimers Dis. 2014, 40, 191–212. [Google Scholar] [CrossRef]
- Sillerud, L.O.; Solberg, N.O.; Chamberlain, R.; Orlando, R.A.; Heidrich, J.E.; Brown, D.C.; Brady, C.I.; Vander Jagt, T.A.; Garwood, M.; Vander Jagt, D.L. SPION-enhanced magnetic resonance imaging of Alzheimer’s disease plaques in AβPP/PS-1 transgenic mouse brain. J. Alzheimers Dis. 2013, 34, 349–365. [Google Scholar] [CrossRef]
- Tafoya, M.A.; Madi, S.; Sillerud, L.O. Superparamagnetic nanoparticle-enhanced MRI of Alzheimer’s disease plaques and activated microglia in 3X transgenic mouse brains: Contrast optimization. J. Magn. Reson. Imaging 2017, 46, 574–588. [Google Scholar] [CrossRef]
- Liu, X.G.; Zhang, L.; Lu, S.; Liu, D.Q.; Zhang, L.X.; Yu, X.L.; Liu, R.T. Multifunctional Superparamagnetic Iron Oxide Nanoparticles Conjugated with Aβ Oligomer-Specific scFv Antibody and Class A Scavenger Receptor Activator Show Early Diagnostic Potentials for Alzheimer’s Disease. Int. J. Nanomed. 2020, 15, 4919–4932. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Guo, X.; Hao, S.; Yang, T.; Wang, J. Iron oxide nanoparticles-loaded hyaluronic acid nanogels for MRI-aided Alzheimer’s disease theranostics. Arab. J. Chem. 2022, 15, 103748. [Google Scholar] [CrossRef]
- Sharma, B.; Grandjean, J.; Phillips, M.; Kumar, A.; Mandino, F.; Yeow, L.Y.; Low, K.J.Y.; Nandwana, V.; Dravid, V.P.; Bengang, X.; et al. Lipocalin-Type Prostaglandin d Synthase Conjugates as Magnetic Resonance Imaging Contrast Agents for Detecting Amyloid β-Rich Regions in the Brain of Live Alzheimer’s Disease Mice. Adv. Nanobiomed Res. 2021, 1, 2100019. [Google Scholar] [CrossRef]
- Gloag, L.; Mehdipour, M.; Ulanova, M.; Mariandry, K.; Nichol, M.A.; Hernández-Castillo, D.J.; Gaudet, J.; Qiao, R.; Zhang, J.; Nelson, M.; et al. Zero valent iron core-iron oxide shell nanoparticles as small magnetic particle imaging tracers. Chem. Commun. 2020, 56, 3504–3507. [Google Scholar] [CrossRef] [PubMed]
- Gloag, L.; Mehdipour, M.; Chen, D.; Tilley, R.D.; Gooding, J.J. Advances in the Application of Magnetic Nanoparticles for Sensing. Adv Mater 2019, 31, 1904385. [Google Scholar] [CrossRef]
- Calero, M.; Gutiérrez, L.; Salas, G.; Luengo, Y.; Lázaro, A.; Acedo, P.; Morales, M.P.; Miranda, R.; Villanueva, A. Efficient and safe internalization of magnetic iron oxide nanoparticles: Two fundamental requirements for biomedical applications. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 733–743. [Google Scholar] [CrossRef]
- Petters, C.; Dringen, R. Accumulation of iron oxide nanoparticles by cultured primary neurons. Neurochem. Int. 2015, 81, 1–9. [Google Scholar] [CrossRef]
- Wierzbinski, K.R.; Szymanski, T.; Rozwadowska, N.; Rybka, J.D.; Zimna, A.; Zalewski, T.; Nowicka-Bauer, K.; Malcher, A.; Nowaczyk, M.; Krupinski, M.; et al. Potential use of superparamagnetic iron oxide nanoparticles for in vitro and in vivo bioimaging of human myoblasts. Sci. Rep. 2018, 8, 3682. [Google Scholar] [CrossRef]
- Silva, L.H.; da Silva, J.R.; Ferreira, G.A.; Silva, R.C.; Lima, E.C.; Azevedo, R.B.; Oliveira, D.M. Labeling mesenchymal cells with DMSA-coated gold and iron oxide nanoparticles: Assessment of biocompatibility and potential applications. J. Nanobiotechnology 2016, 14, 59. [Google Scholar] [CrossRef]
- Wang, C.; Cai, X.; Wang, R.; Zhai, S.; Zhang, Y.; Hu, W.; Zhang, Y.; Wang, D. Neuroprotective effects of verbascoside against Alzheimer’s disease via the relief of endoplasmic reticulum stress in Aβ-exposed U251 cells and APP/PS1 mice. J. Neuroinflammation 2020, 17, 309. [Google Scholar] [CrossRef]
- Luo, R.; Fan, Y.; Yang, J.; Ye, M.; Zhang, D.-F.; Guo, K.; Li, X.; Bi, R.; Xu, M.; Yang, L.-X.; et al. A novel missense variant in ACAA1 contributes to early-onset Alzheimer’s disease, impairs lysosomal function, and facilitates amyloid-β pathology and cognitive decline. Signal Transduct. Target. Ther. 2021, 6, 325. [Google Scholar] [CrossRef]
- Li, Z.; Bi, H.; Jiang, H.; Song, J.; Meng, Q.; Zhang, Y.; Fei, X. Neuroprotective effect of emodin against Alzheimer’s disease via Nrf2 signaling in U251 cells and APP/PS1 mice. Mol. Med. Rep. 2021, 23, 108. [Google Scholar] [CrossRef]
- Kimelberg, H.K.; Nedergaard, M. Functions of astrocytes and their potential as therapeutic targets. Neurotherapeutics 2010, 7, 338–353. [Google Scholar] [CrossRef] [PubMed]
- Geppert, M.; Hohnholt, M.C.; Thiel, K.; Nürnberger, S.; Grunwald, I.; Rezwan, K.; Dringen, R. Uptake of dimercaptosuccinate-coated magnetic iron oxide nanoparticles by cultured brain astrocytes. Nanotechnology 2011, 22, 145101. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.N.; McCann, J.; Yamasaki, E. Methods for detecting carcinogens and mutagens with the salmonella/mammalian-microsome mutagenicity test. Mutat. Res. 1975, 31, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Poljak, A.; Ali, S.A.; Zhong, L.; Raftery, M.J.; Sachdev, P. Extending the Depth of Human Plasma Proteome Coverage Using Simple Fractionation Techniques. J. Proteome Res. 2021, 20, 1261–1279. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Selkoe, D.J. Alzheimer’s disease: Genes, proteins, and therapy. Physiol. Rev. 2001, 81, 741–766. [Google Scholar] [CrossRef]
- Nirmalraj, P.N.; List, J.; Battacharya, S.; Howe, G.; Xu, L.; Thompson, D.; Mayer, M. Complete aggregation pathway of amyloid β (1-40) and (1-42) resolved on an atomically clean interface. Sci. Adv. 2020, 6, eaaz6014. [Google Scholar] [CrossRef]
- Patel, M.M.; Patel, B.M. Crossing the Blood–Brain Barrier: Recent Advances in Drug Delivery to the Brain. CNS Drugs 2017, 31, 109–133. [Google Scholar] [CrossRef]
- Bastrup, J.; Hansen, K.H.; Poulsen, T.B.G.; Kastaniegaard, K.; Asuni, A.A.; Christensen, S.; Belling, D.; Helboe, L.; Stensballe, A.; Volbracht, C. Anti-Aβ Antibody Aducanumab Regulates the Proteome of Senile Plaques and Closely Surrounding Tissue in a Transgenic Mouse Model of Alzheimer’s Disease. J. Alzheimers Dis. 2021, 79, 249–265. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Zhang, J.; Li, X.-Y.; Yuan, L.; Pan, Y.-F.; Chen, X.-R.; Gao, T.-M.; Qiao, J.-T.; Qi, J.-S. A novel antibody targeting sequence 31–35 in amyloid β protein attenuates Alzheimer’s disease-related neuronal damage. Hippocampus 2017, 27, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Widiapradja, A.; Vegh, V.; Lok, K.Z.; Manzanero, S.; Thundyil, J.; Gelderblom, M.; Cheng, Y.-L.; Pavlovski, D.; Tang, S.-C.; Jo, D.-G.; et al. Intravenous immunoglobulin protects neurons against amyloid beta-peptide toxicity and ischemic stroke by attenuating multiple cell death pathways. J. Neurochem. 2012, 122, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Calero, M.; Chiappi, M.; Lazaro-Carrillo, A.; Rodríguez, M.J.; Chichón, F.J.; Crosbie-Staunton, K.; Prina-Mello, A.; Volkov, Y.; Villanueva, A.; Carrascosa, J.L. Characterization of interaction of magnetic nanoparticles with breast cancer cells. J. Nanobiotechnology 2015, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Pudovkin, M.S.; Shamsutdinov, N.I.; Zelenikhin, P.V.; Nizamutdinov, A.S. Transmission electron microscopy and flow cytometry study of cellular uptake of unmodified Pr3+:LaF3 nanoparticles in dynamic. J. Nanopart Res. 2021, 23, 124. [Google Scholar] [CrossRef]
- Portilla, Y.; Mulens-Arias, V.; Daviu, N.; Paradela, A.; Pérez-Yagüe, S.; Barber, D.F. Interaction of Iron Oxide Nanoparticles with Macrophages Is Influenced Distinctly by “Self” and “Non-Self” Biological Identities. ACS Appl. Mater. Interfaces 2023, 15, 35906–35926. [Google Scholar] [CrossRef]
- Kinnear, C.; Moore, T.L.; Rodriguez-Lorenzo, L.; Rothen-Rutishauser, B.; Petri-Fink, A. Form Follows Function: Nanoparticle Shape and Its Implications for Nanomedicine. Chem. Rev. 2017, 117, 11476–11521. [Google Scholar] [CrossRef]
- Wilhelm, C.; Gazeau, F.; Bacri, J.C. Magnetophoresis and ferromagnetic resonance of magnetically labeled cells. Eur. Biophys. J. 2002, 31, 118–125. [Google Scholar] [CrossRef]
- Galimard, A.; Safi, M.; Ould-Moussa, N.; Montero, D.; Conjeaud, H.; Berret, J.F. Thirty-femtogram detection of iron in mammalian cells. Small 2012, 8, 2036–2044. [Google Scholar] [CrossRef]
- Mejías, R.; Pérez-Yagüe, S.; Gutiérrez, L.; Cabrera, L.I.; Spada, R.; Acedo, P.; Serna, C.J.; Lázaro, F.J.; Villanueva, A.; Morales Mdel, P.; et al. Dimercaptosuccinic acid-coated magnetite nanoparticles for magnetically guided in vivo delivery of interferon gamma for cancer immunotherapy. Biomaterials 2011, 32, 2938–2952. [Google Scholar] [CrossRef]
- Villanueva, A.; Cañete, M.; Roca, A.G.; Calero, M.; Veintemillas-Verdaguer, S.; Serna, C.J.; Morales Mdel, P.; Miranda, R. The influence of surface functionalization on the enhanced internalization of magnetic nanoparticles in cancer cells. Nanotechnology 2009, 20, 115103. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Wilhelm, C.; Luciani, N.; Deveaux, V.; Gendron, F.; Luciani, A.; Devaud, M.; Gazeau, F. Nanomagnetism reveals the intracellular clustering of iron oxide nanoparticles in the organism. Nanoscale 2011, 3, 4402–4410. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-R.; Kuo, C.-J.; Lin, H.Y.-H.; Wu, C.-J.; Liang, S.-S. A proteomics analysis to evaluate cytotoxicity in NRK-52E cells caused by unmodified Nano-Fe3O4. Sci. World J. 2014, 2014, 754721. [Google Scholar] [CrossRef] [PubMed]
- Askri, D.; Cunin, V.; Béal, D.; Berthier, S.; Chovelon, B.; Arnaud, J.; Rachidi, W.; Sakly, M.; Amara, S.; Sève, M.; et al. Investigating the toxic effects induced by iron oxide nanoparticles on neuroblastoma cell line: An integrative study combining cytotoxic, genotoxic and proteomic tools. Nanotoxicology 2019, 13, 1021–1040. [Google Scholar] [CrossRef] [PubMed]
- Díez, P.; González-Muñoz, M.; González-González, M.; Dégano, R.M.; Jara-Acevedo, R.; Sánchez-Paradinas, S.; Piñol, R.; Murillo, J.L.; Lou, G.; Palacio, F.; et al. Functional insights into the cellular response triggered by a bile-acid platinum compound conjugated to biocompatible ferric nanoparticles using quantitative proteomic approaches. 10.1039/C7NR02196H. Nanoscale 2017, 9, 9960–9972. [Google Scholar] [CrossRef]
- Hofmann, D.; Tenzer, S.; Bannwarth, M.B.; Messerschmidt, C.; Glaser, S.-F.; Schild, H.; Landfester, K.; Mailänder, V. Mass Spectrometry and Imaging Analysis of Nanoparticle-Containing Vesicles Provide a Mechanistic Insight into Cellular Trafficking. ACS Nano 2014, 8, 10077–10088. [Google Scholar] [CrossRef]
- Luo, T.; Gao, J.; Lin, N.; Wang, J. Effects of Two Kinds of Iron Nanoparticles as Reactive Oxygen Species Inducer and Scavenger on the Transcriptomic Profiles of Two Human Leukemia Cells with Different Stemness. Nanomaterials 2020, 10, 1951. [Google Scholar] [CrossRef]
- Xie, Y.; Jiang, J.; Tang, Q.; Zou, H.; Zhao, X.; Liu, H.; Ma, D.; Cai, C.; Zhou, Y.; Chen, X.; et al. Iron Oxide Nanoparticles as Autophagy Intervention Agents Suppress Hepatoma Growth by Enhancing Tumoricidal Autophagy. Adv. Sci. 2020, 7, 1903323. [Google Scholar] [CrossRef]
- Laskar, A.; Ghosh, M.; Khattak, S.I.; Li, W.; Yuan, X.M. Degradation of superparamagnetic iron oxide nanoparticle-induced ferritin by lysosomal cathepsins and related immune response. Nanomedicine 2012, 7, 705–717. [Google Scholar] [CrossRef]
- Laurent, S.; Burtea, C.; Thirifays, C.; Häfeli, U.O.; Mahmoudi, M. Crucial Ignored Parameters on Nanotoxicology: The Importance of Toxicity Assay Modifications and “Cell Vision”. PLoS ONE 2012, 7, e29997. [Google Scholar] [CrossRef]
- Ngoka, L.C.M. Dramatic down-regulation of oxidoreductases in human hepatocellular carcinoma hepG2 cells: Proteomics and gene ontology unveiling new frontiers in cancer enzymology. Proteome Sci. 2008, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Rauvala, H.; Huttunen, H.J.; Fages, C.; Kaksonen, M.; Kinnunen, T.; Imai, S.; Raulo, E.; Kilpeläinen, I. Heparin-binding proteins HB-GAM (pleiotrophin) and amphoterin in the regulation of cell motility. Matrix Biol. 2000, 19, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.; McFarlane, S.; Mulligan, K.; Gillespie, H.; Draffin, J.E.; Trimble, A.; Ouhtit, A.; Johnston, P.G.; Harkin, D.P.; McCormick, D.; et al. Cortactin underpins CD44-promoted invasion and adhesion of breast cancer cells to bone marrow endothelial cells. Oncogene 2006, 25, 6079–6091. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, M.; Clemons, T.D.; Ho, D.; Gutiérrez, L.; Lázaro, F.J.; House, M.J.; St Pierre, T.G.; Fear, M.W.; Wood, F.M.; Iyer, K.S. Manipulating directional cell motility using intracellular superparamagnetic nanoparticles. Nanoscale 2015, 7, 4884–4889. [Google Scholar] [CrossRef]
- Diana, V.; Bossolasco, P.; Moscatelli, D.; Silani, V.; Cova, L. Dose Dependent Side Effect of Superparamagnetic Iron Oxide Nanoparticle Labeling on Cell Motility in Two Fetal Stem Cell Populations. PLoS ONE 2013, 8, e78435. [Google Scholar] [CrossRef]
- Soenen, S.J.; Himmelreich, U.; Nuytten, N.; Pisanic, T.R., II; Ferrari, A.; De Cuyper, M. Intracellular nanoparticle coating stability determines nanoparticle diagnostics efficacy and cell functionality. Small 2010, 6, 2136–2145. [Google Scholar] [CrossRef]
- Cromer Berman, S.M.; Kshitiz; Wang, C.J.; Orukari, I.; Levchenko, A.; Bulte, J.W.; Walczak, P. Cell motility of neural stem cells is reduced after SPIO-labeling, which is mitigated after exocytosis. Magn. Reson. Med. 2013, 69, 255–262. [Google Scholar] [CrossRef]
- Lim, J.P.; Gleeson, P.A. Macropinocytosis: An endocytic pathway for internalising large gulps. Immunol. Cell Biol. 2011, 89, 836–843. [Google Scholar] [CrossRef]
- Soenen, S.J.; Nuytten, N.; De Meyer, S.F.; De Smedt, S.C.; De Cuyper, M. High intracellular iron oxide nanoparticle concentrations affect cellular cytoskeleton and focal adhesion kinase-mediated signaling. Small 2010, 6, 832–842. [Google Scholar] [CrossRef]
- Vtyurina, N.; Åberg, C.; Salvati, A. Imaging of nanoparticle uptake and kinetics of intracellular trafficking in individual cells. Nanoscale 2021, 13, 10436–10446. [Google Scholar] [CrossRef]
- Kamentseva, R.; Kosheverova, V.; Kharchenko, M.; Zlobina, M.; Salova, A.; Belyaeva, T.; Nikolsky, N.; Kornilova, E. Functional cycle of EEA1-positive early endosome: Direct evidence for pre-existing compartment of degradative pathway. PLoS ONE 2020, 15, e0232532. [Google Scholar] [CrossRef]
- Lakshminarayan, R.; Wunder, C.; Becken, U.; Howes, M.T.; Benzing, C.; Arumugam, S.; Sales, S.; Ariotti, N.; Chambon, V.; Lamaze, C.; et al. Galectin-3 drives glycosphingolipid-dependent biogenesis of clathrin-independent carriers. Nat. Cell Biol. 2014, 16, 595–606. [Google Scholar] [CrossRef]
- Gerber, H.; Mosser, S.; Boury-Jamot, B.; Stumpe, M.; Piersigilli, A.; Goepfert, C.; Dengjel, J.; Albrecht, U.; Magara, F.; Fraering, P.C. The APMAP interactome reveals new modulators of APP processing and beta-amyloid production that are altered in Alzheimer’s disease. Acta Neuropathol. Commun. 2019, 7, 13. [Google Scholar] [CrossRef]
- Al-Khallaf, H. Isocitrate dehydrogenases in physiology and cancer: Biochemical and molecular insight. Cell Biosci. 2017, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Mojsilovic-Petrovic, J.; Andrade, M.F.; Zhang, H.; Ball, M.; Stanimirovic, D.B. The expression and functional characterization of ABCG2 in brain endothelial cells and vessels. Faseb J. 2003, 17, 2085–2087. [Google Scholar] [CrossRef] [PubMed]
- Larsen, R.; Gouveia, Z.; Soares, M.P.; Gozzelino, R. Heme cytotoxicity and the pathogenesis of immune-mediated inflammatory diseases. Front. Pharmacol. 2012, 3, 77. [Google Scholar] [CrossRef]
- Chen, R.; Ling, D.; Zhao, L.; Wang, S.; Liu, Y.; Bai, R.; Baik, S.; Zhao, Y.; Chen, C.; Hyeon, T. Parallel Comparative Studies on Mouse Toxicity of Oxide Nanoparticle- and Gadolinium-Based T1 MRI Contrast Agents. ACS Nano 2015, 9, 12425–12435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Piao, M.J.; Kim, K.C.; Kim, A.D.; Choi, J.Y.; Choi, J.; Hyun, J.W. Endoplasmic reticulum stress signaling is involved in silver nanoparticles-induced apoptosis. Int. J. Biochem. Cell Biol. 2012, 44, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Garantziotis, S. Interplay between apoptotic and autophagy pathways after exposure to cerium dioxide nanoparticles in human monocytes. Autophagy 2013, 9, 101–103. [Google Scholar] [CrossRef]
- Yu, Z.; Zhu, J.; Wang, H.; Li, H.; Jin, X. Function of BCLAF1 in human disease. Oncol. Lett. 2022, 23, 58. [Google Scholar] [CrossRef]
- Kadry, H.; Noorani, B.; Cucullo, L. A blood–brain barrier overview on structure, function, impairment, and biomarkers of integrity. Fluids Barriers CNS 2020, 17, 69. [Google Scholar] [CrossRef] [PubMed]
- Estevanato, L.; Cintra, D.; Baldini, N.; Portilho, F.; Barbosa, L.; Martins, O.; Lacava, B.; Miranda-Vilela, A.L.; Tedesco, A.C.; Báo, S.; et al. Preliminary biocompatibility investigation of magnetic albumin nanosphere designed as a potential versatile drug delivery system. Int. J. Nanomed. 2011, 6, 1709–1717. [Google Scholar] [CrossRef]





| TEM Diameter (nm) | Hydrodynamic Size (nm) | PDI | Zeta Potential (mV) | |
|---|---|---|---|---|
| Iron nanoparticles | 11.6 ± 0.9 | 106.7 | 0.141 | −22.7 ± 6.26 |
| NP-Ab | 369.5 | 0.994 | −31.5 ± 4.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulanova, M.; Gloag, L.; Bongers, A.; Kim, C.-K.; Duong, H.T.K.; Kim, H.N.; Gooding, J.J.; Tilley, R.D.; Biazik, J.; Wen, W.; et al. Evaluation of Dimercaptosuccinic Acid-Coated Iron Nanoparticles Immunotargeted to Amyloid Beta as MRI Contrast Agents for the Diagnosis of Alzheimer’s Disease. Cells 2023, 12, 2279. https://doi.org/10.3390/cells12182279
Ulanova M, Gloag L, Bongers A, Kim C-K, Duong HTK, Kim HN, Gooding JJ, Tilley RD, Biazik J, Wen W, et al. Evaluation of Dimercaptosuccinic Acid-Coated Iron Nanoparticles Immunotargeted to Amyloid Beta as MRI Contrast Agents for the Diagnosis of Alzheimer’s Disease. Cells. 2023; 12(18):2279. https://doi.org/10.3390/cells12182279
Chicago/Turabian StyleUlanova, Marina, Lucy Gloag, Andre Bongers, Chul-Kyu Kim, Hong Thien Kim Duong, Ha Na Kim, John Justin Gooding, Richard D. Tilley, Joanna Biazik, Wei Wen, and et al. 2023. "Evaluation of Dimercaptosuccinic Acid-Coated Iron Nanoparticles Immunotargeted to Amyloid Beta as MRI Contrast Agents for the Diagnosis of Alzheimer’s Disease" Cells 12, no. 18: 2279. https://doi.org/10.3390/cells12182279