Natural Enantiomers: Occurrence, Biogenesis and Biological Properties
Abstract
:1. Introduction
2. Enantiomers from Kingdom Plantae
2.1. Lignans
2.1.1. Acyclic Lignans
2.1.2. Cyclic Lignans
2.1.3. Sesquineolignans
2.2. Coumarins
2.3. Simple Phenylpropanoids
2.4. Alkaloids
2.4.1. Indole Alkaloids
2.4.2. Quinoline and Isoquinoline Alkaloids
2.4.3. β-Carboline and Carbazole Alkaloids
2.4.4. Piperidine Alkaloids
2.4.5. Thiohydantoin Alkaloids
2.4.6. Indolizidine and Quinolizidine Alkaloids
2.4.7. Other Alkaloids
2.5. Flavonoids
2.5.1. Flavones and Isoflavones
2.5.2. Chalcones
2.5.3. Xanthones
2.6. Terpenoids
2.6.1. Sesquiterpenoids
2.6.2. Diterpenoids
2.6.3. Meroterpenoids
2.7. Phloroglucinols
2.8. Naphthalenes and Phenanthrenes
2.8.1. Naphthalenes
2.8.2. Phenanthrenes
2.9. Chromanes
2.10. Acetophenones
2.11. Diarylheptanoids
2.12. Triphenylmethanes
2.13. Fatty Acids
2.14. Miscellaneous
3. Enantiomers from Kingdom Fungi
3.1. Enantiomers from Phylum Ascomycota
3.1.1. Nonalkaloids
3.1.2. Alkaloids
3.2. Enantiomers from Phylum Basidiomycota
4. Enantiomers from Kingdom Prokaryota
5. Enantiomers from Kingdom Animalia
5.1. Enantiomers from Phylum Porifera
5.1.1. Terpenoids
5.1.2. Alkaloids
5.1.3. Lipids
5.2. Enantiomers from Phylum Arthropoda
5.3. Enantiomers from Phylum Chordata
6. Biological Properties
6.1. Cytotoxicity
6.2. Antiviral
6.3. Antibacterial
6.4. Antifungal
6.5. Anti-Inflammation
6.6. Antioxidation
6.7. Cell Protection
6.8. Enzyme Inhibition
6.9. Aβ Aggregation Inhibition
6.10. Miscellaneous Activities
7. Conclusions
7.1. Natural Distribution of Enantiomers
7.2. Natural Formation of Enantiomers
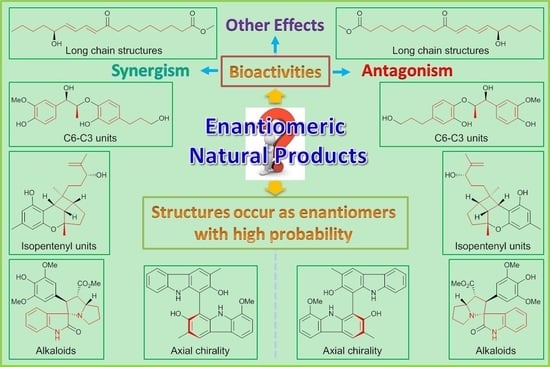
7.3. Structures Tend to Exist as Enantiomers in Nature
7.4. Identification of the Presence of Enantiomers
7.5. Separation and Differentiation of Enantiomers
7.6. Stereochemistry–Bioactivity Relationship of Enantiomers
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dewick, P.M. Medicinal Natural Products: A Biosynthetic Approach, 3rd ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2009; p. 148. [Google Scholar]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Finefield, J.M.; Sherman, D.H.; Kreitman, M.; Williams, R.M. Enantiomeric natural products: Occurrence and biogenesis. Angew. Chem. 2012, 51, 4802–4836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, K. Bioactive natural products and chirality. Chirality 2011, 23, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, K.K.; Gao, X.H.; Yue, J.M. Natural occurrence of all eight stereoisomers of a neosecurinane structure from Flueggea virosa. Tetrahedron 2017, 73, 4692–4697. [Google Scholar] [CrossRef]
- Song, X.Q.; Zhu, K.; Yu, J.H.; Zhang, Q.; Zhang, Y.; He, F.; Cheng, Z.Q.; Jiang, C.S.; Bao, J.; Zhang, H. New octadecanoid enantiomers from the whole plants of Plantago depressa. Molecules 2018, 23, 1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Yu, J.H.; Zhang, J.S.; Song, X.Q.; Bao, J.; Zhang, H. Chromane enantiomers from the flower buds of Tussilago farfara L. and assignments of their absolute configurations. Chem. Biodivers. 2019, 16, e1800581. [Google Scholar] [CrossRef]
- Yu, J.H.; Zhai, H.J.; Yu, Z.P.; Zhang, Q.Q.; Ge, Y.X.; Zhang, Y.Y.; Jiang, C.S.; Zhang, H. Methyl 2-naphthoates from a traditional Chinese herb Morinda officinalis var. officinalis. Tetrahedron 2019, 75, 3793–3801. [Google Scholar] [CrossRef]
- Song, X.Q.; Yu, S.J.; Zhang, J.S.; Yu, J.H.; Zhang, H. New octadecanoid derivatives from the seeds of Ipomoea nil. Chin. J. Nat. Med. 2019, 17, 303–307. [Google Scholar] [CrossRef]
- Batista, A.N.L.; dos Santos, F.M., Jr.; Batista, J.M., Jr.; Cass, Q.B. Enantiomeric mixtures in natural product chemistry: Separation and absolute configuration assignment. Molecules 2018, 23, 492. [Google Scholar] [CrossRef] [Green Version]
- Runeberg, P.A.; Brusentsev, Y.; Rendon, S.M.K.; Eklund, P.C. Oxidative Transformations of Lignans. Molecules 2019, 24, 300. [Google Scholar] [CrossRef] [Green Version]
- Zalesak, F.; Bon, D.J.Y.D.; Pospisil, J. Lignans and Neolignans: Plant secondary metabolites as a reservoir of biologically active substances. Pharmacol. Res. 2019, 146, 104284. [Google Scholar] [CrossRef] [PubMed]
- Moss, G.P. Nomenclature of lignans and neolignans: (IUPAC recommendations 2000). Pure Appl. Chem. 2000, 72, 1493–1523. [Google Scholar] [CrossRef]
- Xu, R.S. Natural product Chemistry, 2nd ed.; Science Press: Beijing, China, 2004. [Google Scholar]
- Liu, X.; Wang, X.B.; Xie, S.S.; Li, Z.R.; Yang, M.H.; Kong, L.Y.; Kim, D.H.; Park, J.S. Lignans from the root of Paeonia lactiflora and their anti-β-amyloid aggregation activities. Fitoterapia 2015, 103, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Xue, Y.; Liu, J.; Yao, G.; Li, D.; Sun, B.; Zhang, J.; Liu, Y.; Qi, C.; Xiang, M.; et al. (±)-Acortatarinowins A-F, Norlignan, Neolignan, and Lignan Enantiomers from Acorus tatarinowii. J. Nat. Prod. 2015, 78, 2205–2214. [Google Scholar] [CrossRef]
- Jiang, C.; Luo, P.; Zhao, Y.; Hong, J.; Morris-Natschke, S.L.; Xu, J.; Chen, C.H.; Lee, K.H.; Gu, Q. Carolignans from the Aerial Parts of Euphorbia sikkimensis and Their Anti-HIV Activity. J. Nat. Prod. 2016, 79, 578–583. [Google Scholar] [CrossRef] [Green Version]
- Seca, A.M.L.; Silva, A.M.S.; Silvestre, A.J.D.; Cavaleiro, J.A.S.; Domingues, F.M.J.; Pascoal-Neto, C. Phenolic constituents from the core of Kenaf (Hibiscus cannabinus). Phytochemistry 2001, 56, 759–767. [Google Scholar] [CrossRef]
- Zhou, L.; Lou, L.L.; Wang, W.; Lin, B.; Chen, J.N.; Wang, X.B.; Huang, X.X.; Song, S.J. Enantiomeric 8-O-4’ type neolignans from red raspberry as potential inhibitors of β-amyloid aggregation. J. Funct. Foods 2017, 37, 322–329. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, H.; Han, F.Y.; Guo, R.; Huang, S.W.; Lin, B.; Huang, X.X.; Song, S.J. Chiral resolution and neuroprotective activities of enantiomeric 8-O-4’ neolignans from the fruits of Crataegus pinnatifida Bge. Fitoterapia 2019, 136, 104164. [Google Scholar] [CrossRef]
- Du, Y.Q.; Yan, Z.Y.; Hou, Z.L.; Guo, R.; Bai, M.; Zhou, L.; Lin, B.; Huang, X.X.; Song, S.J. Enantiomeric 8,4’-type oxyneolignans from the root barks of Ailanthus altissima (Mill.) Swingle and their neuroprotective effects against H2O2-induced SH-SY5Y cells injury. Fitoterapia 2019, 139, 104403. [Google Scholar] [CrossRef]
- Xiong, L.; Zhu, C.; Li, Y.; Tian, Y.; Lin, S.; Yuan, S.; Hu, J.; Hou, Q.; Chen, N.; Yang, Y.; et al. Lignans and neolignans from Sinocalamus affinis and their absolute configurations. J. Nat. Prod. 2011, 74, 1188–1200. [Google Scholar] [CrossRef]
- Braga, A.C.H.; Zacchino, S.; Badano, H.; Gonzalez Sierra, M.; Ruveda, E.A. Carbon-13 NMR spectral and conformational analysis of 8-O-4’ neolignans. Phytochemistry 1984, 23, 2025–2028. [Google Scholar] [CrossRef]
- Gan, M.; Zhang, Y.; Lin, S.; Liu, M.; Song, W.; Zi, J.; Yang, Y.; Fan, X.; Shi, J.; Hu, J.; et al. Glycosides from the root of Iodes cirrhosa. J. Nat. Prod. 2008, 71, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.N.; Han, B.; Yang, P.F.; Feng, Z.M.; Jiang, J.S.; Zhang, P.C. A concise approach for determining the relative configuration of H-7 and H-8 in 8,4′-oxyneolignans by 1H NMR spectroscopy. Org. Chem. Front. 2019, 6, 886–891. [Google Scholar] [CrossRef]
- Lin, S.; Wang, S.; Liu, M.; Gan, M.; Li, S.; Yang, Y.; Wang, Y.; He, W.; Shi, J. Glycosides from the Stem Bark of Fraxinus sieboldiana. J. Nat. Prod. 2007, 70, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Pescitelli, G.; Bruhn, T. Good Computational Practice in the Assignment of Absolute Configurations by TDDFT Calculations of ECD Spectra. Chirality 2016, 28, 466–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, D.-P.; Feng, X.L.; Zhang, W.Y.; Gao, H.; Cheng, X.R.; Zhou, W.X.; Yu, Y.; Yao, X.S. Anti-neuroinflammatory asarone derivatives from the rhizomes of Acorus tatarinowii. RSC Adv. 2017, 7, 8512–8520. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.S.; Cai, L.; Wang, J.P.; Yin, T.P.; Yu, J.; Ding, Z.T.; Fang, Y.S.; Yang, M.H. New phenylpropanoids from Bulbophyllum retusiusculum. Arch. Pharm. Res. 2018, 41, 1074–1081. [Google Scholar] [CrossRef]
- Cheng, Z.B.; Lu, X.; Bao, J.M.; Han, Q.H.; Dong, Z.; Tang, G.H.; Gan, L.S.; Luo, H.B.; Yin, S. (±)-Torreyunlignans A-D, Rare 8-9’ Linked Neolignan Enantiomers as Phosphodiesterase-9A Inhibitors from Torreya yunnanensis. J. Nat. Prod. 2014, 77, 2651–2657. [Google Scholar] [CrossRef]
- Yang, D.T.; Lin, S.S.; Chen, J.H.; Yuan, S.T.; Shi, J.S.; Wang, J.S.; Jia, A.Q. (+)- and (−)-liriodenol, a pair of novel enantiomeric lignans from Liriodendron hybrid. Bioorg. Med. Chem. Lett. 2015, 25, 1976–1978. [Google Scholar] [CrossRef]
- Zhu, Y.; Huang, R.Z.; Wang, C.G.; Ouyang, X.L.; Jing, X.T.; Liang, D.; Wang, H.S. New inhibitors of matrix metalloproteinases 9 (MMP-9): Lignans from Selaginella moellendorffii. Fitoterapia 2018, 130, 281–289. [Google Scholar] [CrossRef]
- Jiao, S.; Su, G.; Zhou, X.; Wuken, S.; Li, J.; Tu, P.; Chai, X. Alashinols I and J, two novel phenols from stem barks of Syringa pinnatifolia. Phytochem. Lett. 2019, 33, 61–63. [Google Scholar] [CrossRef]
- Odonbayar, B.; Murata, T.; Buyankhishig, B.; Sasaki, K.; Suganuma, K.; Ishikawa, Y.; Batkhuu, J. Acylated Lignans Isolated from Brachanthemum gobicum and Their Trypanocidal Activity. J. Nat. Prod. 2019, 82, 774–784. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Komatsu, M.; Kido, M.; Nakagawa, K. Synthesis of lignans. II. A simple biogenetic-type synthesis of magnosalicin, a new neolignan with antiallergy activity isolated from Magnolia salicifolia. Tetrahedron 1986, 42, 523–528. [Google Scholar] [CrossRef]
- Muraoka, O.; Sawada, T.; Morimoto, E.; Tanabe, G. Chalcones as synthetic intermediates. A facile route to (±)-magnosalicin, an antiallergy neolignan. Chem. Pharm. Bull. 1993, 41, 772–774. [Google Scholar] [CrossRef] [Green Version]
- Lopes, N.P.; Blumenthal, E.E.d.A.; Cavalheiro, A.J.; Kato, M.J.; Yoshida, M. Lignans, γ-lactones and propiophenones of Virola surinamensis. Phytochemistry 1996, 43, 1089–1092. [Google Scholar] [CrossRef]
- Lu, Y.; Xue, Y.; Chen, S.; Zhu, H.; Wang, J.; Liu, J.; Qi, C.; Zhang, Y.; Lu, Y.; Zhang, J.; et al. Antioxidant Lignans and Neolignans from Acorus tatarinowii. Sci. Rep. 2016, 6, 22909. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.Y.; Cheng, B.; Zheng, Y.J.; Dong, Z.; Lin, S.L.; Tang, G.H.; Gu, Q.; Yin, S. Enantiomeric neolignans and sesquineolignans from Jatropha integerrima and their absolute configurations. RSC Adv. 2015, 5, 12202–12208. [Google Scholar] [CrossRef]
- Lou, L.L.; Yao, G.D.; Wang, J.; Zhao, W.Y.; Wang, X.B.; Huang, X.X.; Song, S.J. Enantiomeric neolignans from Picrasma quassioides exhibit distinctive cytotoxicity on hepatic carcinoma cells through ROS generation and apoptosis induction. Bioorg. Med. Chem. Lett. 2018, 28, 1263–1268. [Google Scholar] [CrossRef]
- Antus, S.; Kurtan, T.; Juhasz, L.; Kiss, L.; Hollosi, M.; Majer, Z. Chiroptical properties of 2,3-dihydrobenzo[b]furan and chromane chromophores in naturally occurring O-heterocycles. Chirality 2001, 13, 493–506. [Google Scholar] [CrossRef]
- Zhou, L.; Yao, G.D.; Lu, L.W.; Song, X.Y.; Lin, B.; Song, S.J.; Wang, X.B.; Huang, X.X. Neolignans from Red Raspberry (Rubus idaeus L.) Exhibit Enantioselective Neuroprotective Effects against H2O2-Induced Oxidative Injury in SH-SY5Y Cells. J. Agric. Food Chem. 2018, 66, 11390–11397. [Google Scholar] [CrossRef]
- Zhou, L.; Xi, Y.F.; Wang, W.; Lin, B.; Wang, X.B.; Huang, X.X.; Song, S.J. Chiral resolution and bioactivity of enantiomeric benzofuran neolignans from the fruit of Rubus ideaus L. Fitoterapia 2018, 127, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhou, L.; Wu, Y.; Zhu, H.; Hu, Z.; Wang, J.; Luo, Z.; Xue, Y.; Zhang, Y.; Lai, Y.; et al. Enantiomeric Lignans and Neolignans from Phyllanthus glaucus: Enantioseparation and Their Absolute Configurations. Sci. Rep. 2016, 6, 24809. [Google Scholar] [CrossRef] [PubMed]
- Mei, R.Q.; Wang, Y.H.; Du, G.H.; Liu, G.M.; Zhang, L.; Cheng, Y.X. Antioxidant lignans from the fruits of Broussonetia papyrifera. J. Nat. Prod. 2009, 72, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Han, F.Y.; Lu, L.W.; Yao, G.D.; Zhang, Y.Y.; Wang, X.B.; Lin, B.; Huang, X.X.; Song, S.J. Isolation of enantiomeric furolactones and furofurans from Rubus idaeus L. with neuroprotective activities. Phytochemistry 2019, 164, 122–129. [Google Scholar] [CrossRef]
- Li, X.N.; Chu, C.; Tong, S.Q.; Cheng, D.P.; Yan, J.Z. A new furolactone-type lignan from Lycium chinense. Nat. Prod. Res. 2013, 27, 750–752. [Google Scholar] [CrossRef]
- Wang, Y.X.; Lin, B.; Zhou, L.; Yan, Z.Y.; Zhang, H.; Huang, X.X.; Song, S.J. Anti-β-amyloid aggregation activity of enantiomeric furolactone-type lignans from Archidendron clypearia (Jack) I.C.N. Nat. Prod. Res. 2020, 34, 456–463. [Google Scholar] [CrossRef]
- Zhou, X.M.; Zheng, C.J.; Wu, J.T.; Chen, G.Y.; Chen, J.; Sun, C.G. Five new lactone derivatives from the stems of Dendrobium nobile. Fitoterapia 2016, 115, 96–100. [Google Scholar] [CrossRef]
- Liu, W.J.; Chen, Y.J.; Chen, D.N.; Wu, Y.P.; Gao, Y.J.; Li, J.; Zhong, W.J.; Jiang, L. A new pair of enantiomeric lignans from the fruits of Morinda citrifolia and their absolute configuration. Nat. Prod. Res. 2018, 32, 933–938. [Google Scholar] [CrossRef]
- Wang, Y.X.; Zhou, L.; Wang, J.; Lin, B.; Wang, X.B.; Huang, X.X.; Song, S.J. Enantiomeric lignans with anti-β-amyloid aggregation activity from the twigs and leaves of Pithecellobium clypearia Benth. Bioorg. Chem. 2018, 77, 579–585. [Google Scholar] [CrossRef]
- Xi, Y.F.; Liu, S.F.; Hong, W.; Song, X.Y.; Lou, L.L.; Zhou, L.; Yao, G.D.; Lin, B.; Wang, X.B.; Huang, X.X.; et al. Discovery of cycloneolignan enantiomers from Isatis indigotica Fortune with neuroprotective effects against MPP+-induced SH-SY5Y cell injury. Bioorg. Chem. 2019, 88, 102926. [Google Scholar] [CrossRef]
- Fukuda, T.; Nagai, K.; Tomoda, H. (±)-Tylopilusins, Diphenolic Metabolites from the Fruiting Bodies of Tylopilus eximius. J. Nat. Prod. 2012, 75, 2228–2231. [Google Scholar] [CrossRef]
- Lai, Y.; Liu, T.; Sa, R.; Wei, X.; Xue, Y.; Wu, Z.; Luo, Z.; Xiang, M.; Zhang, Y.; Yao, G. Neolignans with a Rare 2-Oxaspiro[4.5]deca-6,9-dien-8-one Motif from the Stem Bark of Cinnamomum subavenium. J. Nat. Prod. 2015, 78, 1740–1744. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, Y.; Li, Y.; Li, L.; Qu, J.; Ma, S.; Yu, S. Chiral resolution and absolute configuration of a pair of rare racemic spirodienone sesquineolignans from Xanthium sibiricum. Org. Lett. 2014, 16, 5406–5409. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Coumarin carbonic anhydrase inhibitors from natural sources. J. Enzyme Inhib. Med. Chem. 2020, 35, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Akkol, E.K.; Genc, Y.; Karpuz, B.; Sobarzo-Sanchez, E.; Capasso, R. Coumarins and Coumarin-Related Compounds in Pharmacotherapy of Cancer. Cancers 2020, 12, 1959. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.J.; Kim, J. Determination of the Absolute Configuration of Khellactone Esters from Peucedanum japonicum Roots. J. Nat. Prod. 2017, 80, 1354–1360. [Google Scholar] [CrossRef] [PubMed]
- Hata, K.; Kozawa, M.; Baba, K.; Yen, K.Y.; Yang, L.L. Coumarins from the roots of Angelica morii. Chem. Pharm. Bull. 1974, 22, 957–961. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.Y.; Wu, F.H.; Wang, J.S.; Li, J.; Kong, L.Y. Attenuation of airway hyperreactivity and T helper cell type 2 responses by coumarins from Peucedanum praeruptorum Dunn in a murine model of allergic airway inflammation. J. Ethnopharmacol. 2012, 141, 314–321. [Google Scholar] [CrossRef]
- Nielsen, B.E.; Larsen, P.K.; Lemmich, J. Constituents of umbelliferous plants. XVII. Coumarins from Seseli gummiferum. Structure of two new coumarins. Acta Chem. Scand. 1971, 25, 529–533. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, H.; Murakami, T.; Nishida, N.; Kageura, T.; Yoshikawa, M. Medicinal foodstuffs. XX. Vasorelaxant active constituents from the roots of Angelica furcijuga Kitagawa: Structures of hyuganins A, B, C, and D. Chem. Pharm. Bull. 2000, 48, 1429–1435. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.L.; Jing, W.H.; Tu, P.F.; Wang, Y.T. Enantiomeric separation of angular-type pyranocoumarins from Peucedani Radix using AD-RH chiral column. Nat. Prod. Res. 2014, 28, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Górski, B.; Talko, A.; Basak, T.; Barbasiewicz, M. Olefination with Sulfonyl Halides and Esters: Scope, Limitations, and Mechanistic Studies of the Hawkins Reaction. Org. Lett. 2017, 19, 1756–1759. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, S.; Lou, H.; Fan, P. New coumarins and monoterpene galloylglycoside from the stem bark of Sapium baccatum. Fitoterapia 2019, 134, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Su, F.; Zhao, Z.; Ma, S.; Wang, R.; Li, Y.; Liu, Y.; Li, L.; Qu, J.; Yu, S.; Li, Y. Cnidimonins A-C, Three Types of Hybrid Dimer from Cnidium monnieri: Structural Elucidation and Semisynthesis. Org. Lett. 2017, 19, 4920–4923. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.H.; Liu, Y.B.; Ma, S.G.; Li, L.; Li, Y.; Jiang, J.D.; Qu, J.; Yu, S.S. Antiviral Spirotriscoumarins A and B: Two Pairs of Oligomeric Coumarin Enantiomers with a Spirodienone-Sesquiterpene Skeleton from Toddalia asiatica. Org. Lett. 2016, 18, 5146–5149. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Tang, G.H.; Guo, F.L.; Li, W.; Zhang, J.S.; Liu, B.; Yin, S. (P)/(M)-corinepalensin A, a pair of axially chiral prenylated bicoumarin enantiomers with a rare C-5-C-5’ linkage from the twigs of Coriaria nepalensis. Phytochemistry 2018, 149, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Lu, S. Biosynthesis and Regulation of Phenylpropanoids in Plants. Crit. Rev. Plant Sci. 2017, 36, 257–290. [Google Scholar] [CrossRef]
- Shi, X.L.; Yan, J.K.; Li, W.K.; Donkor, P.O.; Gao, X.M.; Ding, L.Q.; Qiu, F. Two pairs of phenylpropanoid enantiomers from the leaves of Eucommia ulmoides. J. Asian Nat. Prod. Res. 2018, 20, 1045–1054. [Google Scholar] [CrossRef]
- Baderschneider, B.; Winterhalter, P. Isolation and characterization of novel benzoates, cinnamates, flavonoids, and lignans from Riesling wine and screening for antioxidant activity. J. Agric. Food Chem. 2001, 49, 2788–2798. [Google Scholar] [CrossRef]
- Bai, L.; Wang, Y.; Ge, Y.; Liu, J.; Luan, X. Diastereoselective Synthesis of Dibenzo[b,d]azepines by Pd(II)-Catalyzed [5 + 2] Annulation of o-Arylanilines with Dienes. Org. Lett. 2017, 19, 1734–1737. [Google Scholar] [CrossRef]
- Lu, Y.Y.; Gong, X.P.; Xue, Y.B.; Zhu, H.C.; Li, X.N.; Hu, L.Z.; Guan, J.K.; Zhang, J.W.; Du, G.; Zhang, Y.H. Two pairs of chlorine-containing phenylpropanoid enantiomers from Acorus tatarinowii. Chin. Chem. Lett. 2017, 28, 1460–1464. [Google Scholar] [CrossRef]
- Gao, E.; Zhou, Z.Q.; Zou, J.; Yu, Y.; Feng, X.L.; Chen, G.D.; He, R.R.; Yao, X.S.; Gao, H. Bioactive Asarone-Derived Phenylpropanoids from the Rhizome of Acorus tatarinowii Schott. J. Nat. Prod. 2017, 80, 2923–2929. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yao, G.D.; Song, X.Y.; Wang, J.; Lin, B.; Wang, X.B.; Huang, X.X.; Song, S.J. Neuroprotective Effects of 1,2-Diarylpropane Type Phenylpropanoid Enantiomers from Red Raspberry against H2O2-Induced Oxidative Stress in Human Neuroblastoma SH-SY5Y Cells. J. Agric. Food Chem. 2018, 66, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Rayanil, K.O.; Nimnoun, C.; Tuntiwachwuttikul, P. New phenolics from the wood of Casearia grewiifolia. Phytochem. Lett. 2012, 5, 59–62. [Google Scholar] [CrossRef]
- Yan, J.K.; Shi, X.L.; Donkor, P.O.; Gao, X.M.; Ding, L.Q.; Qiu, F.; Yan, J.K.; Shi, X.L.; Donkor, P.O.; Gao, X.M.; et al. Two pairs of phenolic enantiomers from the leaves of Eucommia ulmoides Oliver. Nat. Prod. Res. 2019, 33, 1162–1168. [Google Scholar] [CrossRef]
- Guo, R.; Shang, X.Y.; Lv, T.M.; Yao, G.D.; Lin, B.; Wang, X.B.; Huang, X.X.; Song, S.J. Phenylpropanoid derivatives from the fruit of Crataegus pinnatifida Bunge and their distinctive effects on human hepatoma cells. Phytochemistry 2019, 164, 252–261. [Google Scholar] [CrossRef]
- Rosales, P.F.; Bordin, G.S.; Gower, A.E.; Moura, S. Indole alkaloids: 2012 until now, highlighting the new chemical structures and biological activities. Fitoterapia 2020, 143, 104558. [Google Scholar] [CrossRef]
- Xu, D.; Xu, Z. Indole Alkaloids with Potential Anticancer Activity. Curr. Top. Med. Chem. 2020, 20, 1938–1949. [Google Scholar] [CrossRef]
- Xi, Y.F.; Lou, L.L.; Xu, Z.Y.; Hou, Z.L.; Wang, X.B.; Huang, X.X.; Song, S.J. Alkaloid Enantiomers from Isatis tinctoria with Neuroprotective Effects against H2O2-Induced SH-SY5Y Cell Injury. Planta Med. 2019, 85, 1374–1382. [Google Scholar] [CrossRef]
- Chen, M.; Gan, L.; Lin, S.; Wang, X.; Li, L.; Li, Y.; Zhu, C.; Wang, Y.; Jiang, B.; Jiang, J.; et al. Alkaloids from the root of Isatis indigotica. J. Nat. Prod. 2012, 75, 1167–1176. [Google Scholar] [CrossRef]
- Liu, J.; Du, Y.Q.; Li, C.J.; Li, L.; Chen, F.Y.; Yang, J.Z.; Chen, N.H.; Zhang, D.M. Alkaloids from the stems of Clausena lansium and their neuroprotective activity. RSC Adv. 2017, 7, 35417–35425. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Deng, A.J.; Li, L.; Wu, L.Q.; Ji, M.; Zhang, H.J.; Li, Z.H.; Ma, L.; Zhang, Z.H.; Chen, X.G.; et al. Azacyclo-indoles and Phenolics from the Flowers of Juglans regia. J. Nat. Prod. 2017, 80, 2189–2198. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.F.; Lin, B.; Xi, Y.F.; Zhou, L.; Lou, L.L.; Huang, X.X.; Wang, X.B.; Song, S.J. Bioactive spiropyrrolizidine oxindole alkaloid enantiomers from Isatis indigotica Fortune. Org. Biomol. Chem. 2018, 16, 9430–9439. [Google Scholar] [CrossRef]
- Zhang, D.; Shi, Y.; Xu, R.; Du, K.; Guo, F.; Chen, K.; Li, Y.; Wang, R. Alkaloid enantiomers from the roots of Isatis indigotica. Molecules 2019, 24, 3140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Q.; Xu, C.; Chen, M.; Lin, S.; Zhu, C.; Jiang, J.; Yang, Y.; Shi, J.; Chen, M.; Li, Y.; et al. Sulfur-enriched alkaloids from the root of Isatis indigotica. Acta Pharm. Sin. B 2018, 8, 933–943. [Google Scholar] [CrossRef]
- Chen, M.; Lin, S.; Li, L.; Zhu, C.; Wang, X.; Wang, Y.; Jiang, B.; Wang, S.; Li, Y.; Jiang, J.; et al. Enantiomers of an Indole Alkaloid Containing Unusual Dihydrothiopyran and 1,2,4-Thiadiazole Rings from the Root of Isatis indigotica. Org. Lett. 2012, 14, 5668–5671. [Google Scholar] [CrossRef]
- Nge, C.E.; Chong, K.W.; Thomas, N.F.; Lim, S.H.; Low, Y.Y.; Kam, T.S. Ibogan, aspidosperman, vincamine, and bisindole alkaloids from a Malayan Tabernaemontana corymbosa: Iboga alkaloids with C-20α substitution. J. Nat. Prod. 2016, 79, 1388–1399. [Google Scholar] [CrossRef]
- Zhang, D.B.; Yu, D.G.; Sun, M.; Zhu, X.X.; Yao, X.J.; Zhou, S.Y.; Chen, J.J.; Gao, K. Ervatamines A-I, Anti-inflammatory Monoterpenoid Indole Alkaloids with Diverse Skeletons from Ervatamia hainanensis. J. Nat. Prod. 2015, 78, 1253–1261. [Google Scholar] [CrossRef]
- Li, D.W.; Guo, Q.L.; Meng, X.H.; Zhu, C.G.; Xu, C.B.; Shi, J.G. Two pairs of unusual scalemic enantiomers from Isatis indigotica leaves. Chin. Chem. Lett. 2016, 27, 1745–1750. [Google Scholar] [CrossRef]
- Li, Y.H.; Zhang, Y.; Peng, L.Y.; Li, X.N.; Zhao, Q.S.; Li, R.T.; Wu, X.D. (±)-Evodiakine, A Pair of Rearranged Rutaecarpine-Type Alkaloids From Evodia rutaecarpa. Nat. Prod. Bioprospect. 2016, 6, 291–296. [Google Scholar] [CrossRef] [Green Version]
- Geng, C.-A.; Huang, X.Y.; Ma, Y.B.; Hou, B.; Li, T.Z.; Zhang, X.M.; Chen, J.J. (±)-Uncarilins A and B, dimeric isoechinulin-type alkaloids from Uncaria rhynchophylla. J. Nat. Prod. 2017, 80, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Dewick, P.M. Medicinal Natural Products: A Biosynthetic Approach, 3rd ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2009; pp. 380–395. [Google Scholar]
- Shang, X.F.; Morris-Natschke, S.L.; Yang, G.Z.; Liu, Y.Q.; Guo, X.; Xu, X.S.; Goto, M.; Li, J.C.; Zhang, J.Y.; Lee, K.H. Biologically active quinoline and quinazoline alkaloids part II. Med. Res. Rev. 2018, 38, 1614–1660. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.F.; Yang, C.J.; MorrisNatschke, S.L.; Li, J.C.; Yin, X.D.; Liu, Y.Q.; Guo, X.; Peng, J.W.; Goto, M.; Zhang, J.Y.; et al. Biologically active isoquinoline alkaloids covering 2014-2018. Med. Res. Rev. 2020, 40, 2212–2289. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.N.; Guo, X.X.; Liu, S.; Feng, L.; Bi, Q.R.; Wang, Z.; Tan, N.H. (±)-Zanthonitidine A, a Pair of Enantiomeric Furoquinoline Alkaloids from Zanthoxylum nitidum with Antibacterial Activity. Nat. Prod. Bioprospect. 2018, 8, 361–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Wang, X.; Lin, S.; Li, L.; Shi, J.; Chen, M. Three pairs of alkaloid enantiomers from the root of Isatis indigotica. Acta Pharm. Sin. B 2016, 6, 141–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, X.; Bai, R.; Guo, Q.; Su, G.; Wang, J.; Yang, X.; Li, L.; Tu, P.; Chai, X. Hendersine A, a novel isoquinoline alkaloid from Corydalis hendersonii. Tetrahedron Lett. 2016, 57, 4858–4862. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Q.Y.; Tu, P.F.; Liang, H.; Xu, F.C. Mucroniferanines A-G, Isoquinoline Alkaloids from Corydalis mucronifera. J. Nat. Prod. 2018, 81, 364–370. [Google Scholar] [CrossRef]
- Feng, F.; Ye, F.Z.; Li, C.L.; Liu, W.Y.; Xie, N. Two new benzo phenanthridine isoquinoline alkaloids from Macleaya cordata. Chin. J. Nat. Med. 2012, 10, 378–382. [Google Scholar] [CrossRef]
- Sai, C.M.; Li, D.H.; Li, S.G.; Han, T.; Guo, Y.Z.; Pei, Y.H.; Bai, J.; Jing, Y.K.; Li, Z.L.; Hua, H.M. Racemic alkaloids from Macleaya cordata: Structural elucidation, chiral resolution, and cytotoxic, antibacterial activities. RSC Adv. 2016, 6, 41173–41180. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, Z.; Han, N.; Jiang, B.; Guo, D.; Teng, F.; Wang, Y.; Yin, J. Ambidalmines A-E and ambidimerine F: Bioactive dihydrobenzophenanthridine alkaloids from Corydalis ambigua var. amurensis. Eur. J. Med. Chem. 2014, 84, 417–424. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, S.Y.; Chen, L.; Huang, X.J.; Zhang, Q.W.; Jiang, R.W.; Yao, F.; Ye, W.C. New enantiomeric isoquinoline alkaloids from Coptis chinensis. Phytochem. Lett. 2014, 7, 89–92. [Google Scholar] [CrossRef]
- Sai, C.M.; Li, D.H.; Xue, C.M.; Wang, K.B.; Hu, P.; Pei, Y.H.; Bai, J.; Jing, Y.K.; Li, Z.L.; Hua, H.M. Two Pairs of Enantiomeric Alkaloid Dimers from Macleaya cordata. Org. Lett. 2015, 17, 4102–4105. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Dan, W.; Schneider, U.; Wang, J. beta-Carboline alkaloid monomers and dimers: Occurrence, structural diversity, and biological activities. Eur. J. Med. Chem. 2018, 157, 622–656. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Feng, Z.L.; Wang, Y.T.; Lin, L.G. Anticancer carbazole alkaloids and coumarins from Clausena plants: A review. Chin. J. Nat. Med. 2017, 15, 881–888. [Google Scholar] [CrossRef]
- Zhao, W.Y.; Zhou, W.Y.; Chen, J.J.; Yao, G.D.; Lin, B.; Wang, X.B.; Huang, X.X.; Song, S.J. Enantiomeric β-carboline dimers from Picrasma quassioides and their anti-hepatoma potential. Phytochemistry 2019, 159, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.M.; Li, F.; Zheng, F.F.; Gong, N.N.; Li, Y.; Feng, W.Z.; Tian, L. (±)-Quassidine K, a pair of cytotoxic bis-β-carboline alkaloid enantiomers from Picrasma quassioides. Nat. Prod. Res. 2020, 34, 489–493. [Google Scholar] [CrossRef]
- Jiao, W.H.; Chen, G.D.; Gao, H.; Li, J.; Gu, B.B.; Xu, T.T.; Yu, H.B.; Shi, G.H.; Yang, F.; Yao, X.S.; et al. (±)-Quassidines I and J, two pairs of cytotoxic bis-β-carboline alkaloid enantiomers from Picrasma quassioides. J. Nat. Prod. 2014, 77, 2707–2712. [Google Scholar] [CrossRef]
- Wang, K.B.; Li, S.G.; Huang, X.Y.; Li, D.H.; Li, Z.L.; Hua, H.M. (±)-Peharmaline A: A Pair of Rare β-Carboline-Vasicinone Hybrid Alkaloid Enantiomers from Peganum harmala. Eur. J. Org. Chem. 2017, 2017, 1876–1879. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, H.Y.; Xu, H.Z.; Liu, J.J.; Meng, X.G.; Zhou, M.; Ruan, H.L. Alkaloids with Immunosuppressive Activity from the Bark of Pausinystalia yohimbe. J. Nat. Prod. 2018, 81, 1841–1849. [Google Scholar] [CrossRef]
- Cao, N.; Chen, Y.; Ma, X.; Zeng, K.; Zhao, M.; Tu, P.; Li, J.; Jiang, Y. Bioactive carbazole and quinoline alkaloids from Clausena dunniana. Phytochemistry 2018, 151, 1–8. [Google Scholar] [CrossRef]
- Ma, X.; Cao, N.; Zhang, C.; Guo, X.; Zhao, M.; Tu, P.; Jiang, Y. Cytotoxic carbazole alkaloid derivatives from the leaves and stems of Murraya microphylla. Fitoterapia 2018, 127, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.L.; Li, J.; Zheng, J.; Gu, X.P.; Ferreira, D.; Zjawiony, J.K.; Zhao, M.B.; Guo, X.Y.; Tu, P.F.; Jiang, Y. LC-MS-guided isolation of insulin-secretion-promoting monoterpenoid carbazole alkaloids from Murraya microphylla. J. Nat. Prod. 2018, 81, 2371–2380. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.B.; Gao, J.; Zou, G.A.; Xin, X.L.; Aisa, H.A. Piperidine Alkaloids with Diverse Skeletons from Anacyclus pyrethrum. J. Nat. Prod. 2018, 81, 1474–1482. [Google Scholar] [CrossRef]
- Chen, Q.B.; Aisa, H.A. Alkaloid constituents from Viola tianschanica. Phytochemistry 2017, 144, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Song, W.W.; Zeng, G.Z.; Peng, W.W.; Chen, K.X.; Tan, N.H. Cytotoxic amides and quinolones from Clausena lansium. Helv. Chim. Acta 2014, 97, 298–305. [Google Scholar] [CrossRef]
- Yu, M.Y.; Qin, X.J.; Peng, X.R.; Wang, X.; Tian, X.X.; Li, Z.R.; Qiu, M.H. Macathiohydantoins B-K, novel thiohydantoin derivatives from Lepidium meyenii. Tetrahedron 2017, 73, 4392–4397. [Google Scholar] [CrossRef]
- Sundaram, G.S.M.; Venkatesh, C.; Ila, H.; Junjappa, H. 1-(Methyldithiocarbonyl)imidazole as thiocarbonyl transfer reagent: A facile one-pot three-component synthesis of 3,5- and 1,3,5-substituted 2-thiohydantoins. Synlett 2007, 2, 251–254. [Google Scholar] [CrossRef]
- Al-Khdhairawi, A.A.Q.; Krishnan, P.; Lim, K.H.; Mai, C.W.; Leong, C.O.; Chung, F.F.L.; Leong, C.O.; Yong, K.T.; Chong, K.W.; Low, Y.Y.; et al. A Bis-benzopyrroloisoquinoline Alkaloid Incorporating a Cyclobutane Core and a Chlorophenanthroindolizidine Alkaloid with Cytotoxic Activity from Ficus fistulosa var. tengerensis. J. Nat. Prod. 2017, 80, 2734–2740. [Google Scholar] [CrossRef]
- Ratnagiriswaran, A.N.; Venkatachalam, K. The chemical examination of Tylophora asthmatica and the isolation of the alkaloids tylophorine and tylophorinine. Indian J. Med. Res. 1935, 22, 433–441. [Google Scholar]
- Stoye, A.; Peez, T.E.; Opatz, T. Left, Right, or Both? On the Configuration of the Phenanthroindolizidine Alkaloid Tylophorine from Tylophora indica. J. Nat. Prod. 2013, 76, 275–278. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, C.; Zhao, X.; Wang, Y.; Feng, D.; Zhang, M.; Xie, H. (±)-Homocrepidine A, a Pair of Anti-inflammatory Enantiomeric Octahydroindolizine Alkaloid Dimers from Dendrobium crepidatum. J. Nat. Prod. 2016, 79, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Li, S.G.; Wang, K.B.; Gong, C.; Bao, Y.; Qin, N.B.; Li, D.H.; Li, Z.L.; Bai, J.; Hua, H.M. Cytotoxic quinazoline alkaloids from the seeds of Peganum harmala. Bioorg. Med. Chem. Lett. 2018, 28, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.D.; Cheng, X.M.; Liu, W.; Han, Z.Z.; Chou, G.X.; Wang, Y.; Sun, D.X.; Wang, Z.T.; Wang, C.H. Peganumine B-I and two enantiomers: New alkaloids from the seeds of Peganum harmala Linn. and their potential cytotoxicity and cholinesterase inhibitory activities. RSC Adv. 2016, 6, 15976–15987. [Google Scholar] [CrossRef]
- Shou, Q.; Banbury, L.K.; Renshaw, D.E.; Smith, J.E.; He, X.; Dowell, A.; Griesser, H.J.; Heinrich, M.; Wohlmuth, H. Parvifloranines A and B, Two 11-Carbon Alkaloids from Geijera parviflora. J. Nat. Prod. 2013, 76, 1384–1387. [Google Scholar] [CrossRef]
- Xia, G.Y.; Owusu, D.P.; Ding, L.Q.; Qiu, F.; Xia, G.Y.; Sun, D.J.; Ma, J.H.; Liu, Y.; Chen, L.X.; Zhao, F. (+)/(−)-Phaeocaulin A-D, four pairs of new enantiomeric germacrane-type sesquiterpenes from Curcuma phaeocaulis as natural nitric oxide inhibitors. Sci. Rep. 2017, 7, 43576. [Google Scholar] [CrossRef]
- Han, Q.T.; Li, G.S.; Xiang, K.L.; Ren, Y.; Dai, S.J. Flavonoid alkaloids from Scutellaria moniliorrhiza with anti-inflammatory activities and inhibitory activities against aldose reductase. Phytochemistry 2018, 152, 91–96. [Google Scholar] [CrossRef]
- Zhou, D.; Chen, G.; Ma, Y.P.; Wang, C.G.; Lin, B.; Yang, Y.Q.; Li, W.; Koike, K.; Hou, Y.; Li, N. Isolation, structural elucidation, optical resolution, and antineuroinflammatory activity of phenanthrene and 9,10-dihydrophenanthrene derivatives from Bletilla striata. J. Nat. Prod. 2019, 82, 2238–2245. [Google Scholar] [CrossRef]
- Cheng, Z.Y.; Du, Y.Q.; Zhang, Q.; Lin, B.; Gao, P.Y.; Huang, X.X.; Song, S.J. Two pairs of new alkaloid enantiomers with a spiro [benzofuranone-benzazepine] skeleton from the bark of Juglans mandshurica. Tetrahedron Lett. 2018, 59, 2050–2053. [Google Scholar] [CrossRef]
- Chen, F.; Huang, X.J.; Liang, Q.P.; Huang, Y.P.; Lan, T.; Zhou, G.X. Three new lignanamides from the root of Lycium chinense with anti-inflammatory activity. Nat. Prod. Res. 2019, 33, 3378–3382. [Google Scholar] [CrossRef]
- Zhu, G.Y.; Yang, J.; Yao, X.J.; Yang, X.; Fu, J.; Liu, X.; Bai, L.P.; Liu, L.; Jiang, Z.H. (±)-Sativamides A and B, two pairs of racemic nor-lignanamide enantiomers from the fruits of Cannabis sativa. J. Org. Chem. 2018, 83, 2376–2381. [Google Scholar] [CrossRef]
- Azmi, M.N.; Chan, G.; Peresse, T.; Remeur, C.; Roussi, F.; Litaudon, M.; Awang, K. Kingianins O-Q: Pentacyclic polyketides from Endiandra kingiana as inhibitor of Mcl-1/Bid interaction. Fitoterapia 2016, 109, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Song, J.; Liu, A.; Xiao, B.; Li, S.; Wen, Z.; Lu, Y.; Du, G. Research Progress of the Antiviral Bioactivities of Natural Flavonoids. Nat. Prod. Bioprospect. 2020, 10, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Xu, P.S.; Xu, K.P.; Zou, Z.X.; Zhou, G.; Li, D.; Li, D.; Li, X.M.; Li, J.; Tan, G.S. Two new biflavanoids from Selaginella trichoclada Alsto. Nat. Prod. Res. 2021, 35, 3410–3416. [Google Scholar] [CrossRef] [PubMed]
- Gaffield, W. Circular dichroism, optical rotatory dispersion, and absolute configuration of flavanones, 3-hydroxyflavanones, and their glycosides. Determination of aglycone chirality in flavanone glycosides. Tetrahedron 1970, 26, 4093–4108. [Google Scholar] [CrossRef]
- Zhang, T.Y.; Lin, J.B.; Li, Q.Z.; Kang, J.C.; Pan, J.L.; Hou, S.H.; Chen, C.; Zhang, S.Y. Copper-Catalyzed Selective ortho-C–H/N–H Annulation of Benzamides with Arynes: Synthesis of Phenanthridinone Alkaloids. Org. Lett. 2017, 19, 1764–1767. [Google Scholar] [CrossRef]
- Li, R.; Cheng, J.; Jiao, M.; Guo, C.; Chen, S.; Li, L.; Liu, A. New phenylpropanoid-substituted flavan-3-ols and flavonols from the leaves of Uncaria rhynchophylla. Fitoterapia 2017, 116, 17–23. [Google Scholar] [CrossRef]
- Zaki, M.A.; Hetta, M.H.; Mohammed, R.; Nanayakkara, N.P.D.; Jacob, M.R.; Khan, S.I.; Ibrahim, M.A.; Samoylenko, V.; Coleman, C.; Ferreira, D.; et al. Bioactive Formylated Flavonoids from Eugenia rigida: Isolation, Synthesis, and X-ray Crystallography. J. Nat. Prod. 2016, 79, 2341–2349. [Google Scholar] [CrossRef]
- Li, Y.; Qin, X.B.; Liu, H.X.; Xu, Z.F.; Tan, H.B.; Qiu, S.X. Two pairs of enantiomeric propylated flavonoids and a new lignan from the aerial parts of Abrus precatorius. Fitoterapia 2019, 133, 125–129. [Google Scholar] [CrossRef]
- He, Q.F.; Wu, Z.L.; Huang, X.J.; Zhong, Y.L.; Jiang, R.W.; Li, Y.L.; Ye, W.C.; Wang, Y.; He, Q.-F.; Wu, Z.L.; et al. Cajanusflavanols A-C, Three Pairs of Flavonostilbene Enantiomers from Cajanus cajan. Org. Lett. 2018, 20, 876–879. [Google Scholar] [CrossRef]
- Xu, L.; Huang, T.; Huang, C.; Wu, C.; Jia, A.; Hu, X. Chiral separation, absolute configuration, and bioactivity of two pairs of flavonoid enantiomers from Morus nigra. Phytochemistry 2019, 163, 33–37. [Google Scholar] [CrossRef]
- Xu, L.J.; Huang, C.Y.; Niu, L.X.; Wu, C.Z.; Yang, P.M.; Yu, M.H.; Wang, Y.F.; Hu, X. Isoprenylated flavonoids from Morus nigra and their PPAR γ agonistic activities. Fitoterapia 2018, 127, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.Q.; Liu, Q.X.; Li, H.L.; Han, W. Flavane constituents from branch of Celastrus hindsii. Zhongcaoyao 2014, 45, 2132–2135. [Google Scholar]
- Li, F.F.; Sun, Q.; Wang, D.; Liu, S.; Lin, B.; Liu, C.T.; Li, L.Z.; Huang, X.X.; Song, S.J. Chiral Separation of Cytotoxic Flavan Derivatives from Daphne giraldii. J. Nat. Prod. 2016, 79, 2236–2242. [Google Scholar] [CrossRef] [PubMed]
- Pang, D.R.; Su, X.Q.; Sun, J.; Li, Y.T.; Zhu, Z.X.; Song, Y.L.; Zhao, Y.F.; Tu, P.F.; Zheng, J.; Li, J. Flavonoid dimers from the total phenolic extract of Chinese dragon’s blood, the red resin of Dracaena cochinchinensis. Fitoterapia 2016, 115, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Hiep, N.T.; Lee, D.; Hiep, N.T.; Kwon, J.; Hong, S.; Mar, W.; Kim, N.; Guo, Y.; Hwang, B.Y. Enantiomeric Isoflavones with neuroprotective activities from the Fruits of Maclura tricuspidata. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Liu, Z.; Zheng, X.; Wang, Y.; Tang, M.; Chen, S.; Zhang, F.; Li, L.; Zhang, C.; Sun, Y. Lignans and isoflavonoids from the stems of Pisonia umbellifera. RSC Adv. 2018, 8, 16383–16391. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Wu, P.; Shen, F.; Ji, J.; Rakesh, K.P. Chalcone derivatives and their antibacterial activities: Current development. Bioorg. Chem. 2019, 91, 103133. [Google Scholar] [CrossRef]
- Shi, Y.S.; Hu, W.Z.; Zhang, X.F.; Lv, X.; Shi, Y.S.; Zhang, Y.; Fu, X. Dihydrochalcones and Diterpenoids from Pteris ensiformis and Their Bioactivities. Molecules 2017, 22, 1413. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, X.; Kelsang, N.; Tu, G.; Kong, D.; Lu, J.; Zhang, Y.; Liang, H.; Tu, P.; Zhang, Q. Structurally diverse cytotoxic dimeric chalcones from Oxytropis chiliophylla. J. Nat. Prod. 2018, 81, 307–315. [Google Scholar] [CrossRef]
- Simard, F.; Gauthier, C.; Chiasson, E.; Lavoie, S.; Mshvildadze, V.; Legault, J.; Pichette, A. Antibacterial balsacones J-M, hydroxycinnamoylated dihydrochalcones from Populus balsamifera buds. J. Nat. Prod. 2015, 78, 1147–1153. [Google Scholar] [CrossRef]
- Ma, Q.; Min, K.; Li, H.L.; Jiang, J.H.; Liu, Y.; Zhan, R.; Chen, Y.G. Horsfiequinones A-F, Dimeric Diarylpropanoids from Horsfieldia tetratepala. Planta Med. 2014, 80, 688–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Pogam, P.; Boustie, J. Xanthones of Lichen Source: A 2016 Update. Molecules 2016, 21, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, C.; Gong, C.; Pu, J.; Li, D.; Li, Z.; Hua, H.; Jia, C.; Jia, C.; Gong, C.; Pu, J.; et al. A pair of new enantiomers of xanthones from the stems and leaves of Cratoxylum cochinchinense. Chin. Med. 2019, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macabeo, A.P.G.; Martinez, F.P.A.; Kurtan, T.; Toth, L.; Mandi, A.; Schmidt, S.; Heilmann, J.; Alejandro, G.J.D.; Knorn, M.; Dahse, H.-M.; et al. Tetrahydroxanthene-1,3(2H)-dione derivatives from Uvaria valderramensis. J. Nat. Prod. 2014, 77, 2717–2721. [Google Scholar] [CrossRef] [PubMed]
- Boonnak, N.; Chantrapromma, S.; Fun, H.K.; Yuenyongsawad, S.; Patrick, B.O.; Maneerat, W.; Williams, D.E.; Andersen, R.J. Three types of cytotoxic natural caged-scaffolds: Pure enantiomers or partial racemates. J. Nat. Prod. 2014, 77, 1562–1571. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.L.; Li, D.H.; Li, X.Y.; Wang, Y.T.; Li, S.G.; Bai, J.; Pei, Y.H.; Jing, Y.K.; Li, Z.L.; Hua, H.M. Bioassay- and chemistry-guided isolation of scalemic caged prenylxanthones from the leaves of Garcinia bracteata. J. Nat. Prod. 2018, 81, 749–757. [Google Scholar] [CrossRef]
- Sriyatep, T.; Andersen, R.J.; Patrick, B.O.; Pyne, S.G.; Muanprasat, C.; Seemakhan, S.; Borwornpinyo, S.; Laphookhieo, S. Scalemic Caged Xanthones Isolated from the Stem Bark Extract of Garcinia propinqua. J. Nat. Prod. 2017, 80, 1658–1667. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.F.; Liu, Z.P.; Wang, F.P. Natural Sesquiterpenoids as Cytotoxic Anticancer Agents. Mini-Rev. Med. Chem. 2011, 11, 1153–1164. [Google Scholar] [CrossRef]
- Xi, F.M.; Ma, S.-G.; Liu, Y.-B.; Li, L.; Yu, S.-S. Artaboterpenoids A and B, Bisabolene-Derived Sesquiterpenoids from Artabotrys hexapetalus. Org. Lett. 2016, 18, 3374–3377. [Google Scholar] [CrossRef]
- Yan, J.; Shi, X.; Donkor, P.O.; Qiu, F.; Yan, J.; Shi, X.; Donkor, P.O.; Gao, X.; Ding, L.; Qiu, F.; et al. Nine pairs of megastigmane enantiomers from the leaves of Eucommia ulmoides Oliver. J. Nat. Med. 2017, 71, 780–790. [Google Scholar] [CrossRef]
- Zhang, R.; Feng, X.; Su, G.; Mu, Z.; Zhang, H.; Zhao, Y.; Jiao, S.; Cao, L.; Chen, S.; Tu, P.; et al. Bioactive Sesquiterpenoids from the Peeled Stems of Syringa pinnatifolia. J. Nat. Prod. 2018, 81, 1711–1720. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.Y.; Zhang, J.L.; Ge, C.Y.; Zhang, J.L. Bioactive sesquiterpenoids and steroids from the resinous exudates of Commiphora myrrha. Nat. Prod. Res. 2019, 33, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Y.; Song, Y.N.; Zhang, L.J.; Zhang, M.; Ye, Y.; Zhang, H.; Zhu, J.Y.; Luo, L.; Xia, J.; Rahman, K. Sesquiterpenes and lignans from the flower buds of Daphne genkwa and their nitric oxide inhibitory activities. Nat. Prod. Res. 2018, 32, 2893–2899. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Pu, J.X.; Du, X.; Li, X.N.; Sun, H.D. Two new guaianolide-type sesquiterpenoids from Kadsura interior. Chin. Chem. Lett. 2013, 24, 111–113. [Google Scholar] [CrossRef]
- Li, H.; Jiao, R.; Mu, J.; Xu, S.; Li, X.; Wang, X.; Li, Z.; Xu, J.; Hua, H.; Li, D. Bioactive Natural Spirolactone-Type 6,7-seco-ent-Kaurane Diterpenoids and Synthetic Derivatives. Molecules 2018, 23, 2914. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.Y.; Gao, K.; Yue, J.M.; Zhao, J.X.; Sheng, L.; Fan, Y.Y.; Li, J.Y.; Yue, J.M. Mangelonoids A and B, Two Pairs of Macrocyclic Diterpenoid Enantiomers from Croton mangelong. Org. Lett. 2018, 20, 4040–4043. [Google Scholar] [CrossRef]
- Feng, L.; Mandi, A.; Tang, C.; Kurtan, T.; Tang, S.; Ke, C.Q.; Shen, N.; Lin, G.; Yao, S.; Ye, Y. A Pair of Enantiomeric Bis-seco-abietane Diterpenoids from Cryptomeria fortunei. J. Nat. Prod. 2018, 81, 2667–2672. [Google Scholar] [CrossRef]
- Jiang, Y.J.; Zhang, Y.; He, J.; Wu, X.D.; Shao, L.D.; Li, X.N.; Su, J.; Peng, L.Y.; Li, R.T.; Zhao, Q.-S. (±)-Salviaprione, a pair of unprecedented abietane-type diterpenoids from Salvia prionitis. Tetrahedron Lett. 2015, 56, 5457–5459. [Google Scholar] [CrossRef]
- Geng, C.A.; Chen, X.L.; Zhou, N.J.; Chen, H.; Ma, Y.B.; Huang, X.Y.; Zhang, X.M.; Chen, J.J. LC-MS Guided Isolation of (±)-Sweriledugenin A, a Pair of Enantiomeric Lactones from Swertia leducii. Org. Lett. 2014, 16, 370–373. [Google Scholar] [CrossRef]
- Liang, W.J.; Geng, C.A.; Zhang, X.M.; Chen, H.; Yang, C.Y.; Rong, G.Q.; Zhao, Y.; Xu, H.B.; Wang, H.; Zhou, N.J.; et al. (±)-Paeoveitol, a Pair of New Norditerpene Enantiomers from Paeonia veitchii. Org. Lett. 2014, 16, 424–427. [Google Scholar] [CrossRef]
- Hu, L.; Zhu, H.; Li, L.; Huang, J.; Sun, W.; Liu, J.; Li, H.; Luo, Z.; Wang, J.; Xue, Y.; et al. (±)-Japonones A and B, two pairs of new enantiomers with anti-KSHV activities from Hypericum japonicum. Sci. Rep. 2016, 6, 27588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, N.; Kobayashi, J.i. Prenylated acylphloroglucinols and meroterpenoids from Hypericum plants. Heterocycles 2015, 90, 23–40. [Google Scholar]
- Huang, G.H.; Hu, Z.; Lei, C.; Wang, P.-P.; Yang, J.; Li, J.Y.; Li, J.; Hou, A.J. Enantiomeric Pairs of Meroterpenoids with Diverse Heterocyclic Systems from Rhododendron nyingchiense. J. Nat. Prod. 2018, 81, 1810–1818. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.B.; Huang, G.H.; Yu, M.H.; Lei, C.; Hou, A.J. Five Pairs of Meroterpenoid Enantiomers from Rhododendron capitatum. J. Org. Chem. 2017, 82, 1632–1637. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.B.; Lei, C.; Gao, L.X.; Li, J.Y.; Li, J.; Hou, A.J. Two Enantiomeric Pairs of Meroterpenoids from Rhododendron capitatum. Org. Lett. 2015, 17, 5040–5043. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, L.; Zhu, R.; Zhang, J.; Zhou, J.; Lou, H. Bibenzyl-based meroterpenoid enantiomers from the Chinese liverwort Radula sumatrana. J. Nat. Prod. 2017, 80, 3143–3150. [Google Scholar] [CrossRef]
- Li, C.; Li, C.J.; Ma, J.; Huang, J.W.; Wang, X.Y.; Wang, X.-L.; Ye, F.; Zhang, D.M. Magmenthanes A-H: Eight new meroterpenoids from the bark of Magnolia officinalis var. Biloba. Bioorg. Chem. 2019, 88, 102948. [Google Scholar] [CrossRef]
- Bridi, H.; Meirelles, G.d.C.; von Poser, G.L. Structural diversity and biological activities of phloroglucinol derivatives from Hypericum species. Phytochemistry 2018, 155, 203–232. [Google Scholar] [CrossRef]
- Cheng, M.J.; Yang, X.Y.; Cao, J.Q.; Liu, C.; Zhong, L.P.; Wang, Y.; You, X.F.; Li, C.C.; Wang, L.; Ye, W.C. Isolation, structure elucidation, and total synthesis of myrtuspirone A from Myrtus communis. Org. Lett. 2019, 21, 1583–1587. [Google Scholar] [CrossRef]
- Tantapakul, C.; Maneerat, W.; Sripisut, T.; Ritthiwigrom, T.; Andersen, R.J.; Cheng, P.; Cheenpracha, S.; Raksat, A.; Laphookhieo, S. New Benzophenones and Xanthones from Cratoxylum sumatranum ssp. neriifolium and Their Antibacterial and Antioxidant Activities. J. Agric. Food Chem. 2016, 64, 8755–8762. [Google Scholar] [CrossRef]
- Hu, L.; Xue, Y.; Zhang, J.; Zhu, H.; Chen, C.; Li, X.-N.; Liu, J.; Wang, Z.; Zhang, Y.; Zhang, Y. (±)-Japonicols A-D, Acylphloroglucinol-Based Meroterpenoid Enantiomers with Anti-KSHV Activities from Hypericum japonicum. J. Nat. Prod. 2016, 79, 1322–1328. [Google Scholar] [CrossRef] [PubMed]
- Cheenpracha, S.; Pyne, S.G.; Patrick, B.O.; Andersen, R.J.; Maneerat, W.; Laphookhieo, S. Mallopenins A-E, antibacterial phenolic derivatives from the fruits of Mallotus philippensis. J. Nat. Prod. 2019, 82, 2174–2180. [Google Scholar] [CrossRef] [PubMed]
- Hans, M.; Charpentier, M.; Huch, V.; Jauch, J.; Bruhn, T.; Bringmann, G.; Quandt, D. Stereoisomeric Composition of Natural Myrtucommulone A. J. Nat. Prod. 2015, 78, 2381–2389. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Tian, H.Y.; Huang, X.L.; Wang, W.J.; Li, N.P.; He, J.; Ye, W.C.; Wang, L. Xanthchrysones A-C: Rearranged Phenylpropanoyl-Phloroglucinol Dimers with Unusual Skeletons from Xanthostemon chrysanthus. J. Org. Chem. 2019, 84, 15355–15361. [Google Scholar] [CrossRef] [PubMed]
- Su, J.C.; Wang, S.; Cheng, W.; Huang, X.J.; Li, M.M.; Jiang, R.W.; Li, Y.L.; Wang, L.; Ye, W.C.; Wang, Y. Phloroglucinol Derivatives with Unusual Skeletons from Cleistocalyx operculatus and Their in Vitro Antiviral Activity. J. Org. Chem. 2018, 83, 8522–8532. [Google Scholar] [CrossRef] [PubMed]
- Oya, A.; Tanaka, N.; Kusama, T.; Kim, S.Y.; Hayashi, S.; Kojoma, M.; Hishida, A.; Kawahara, N.; Sakai, K.; Gonoi, T.; et al. Prenylated Benzophenones from Triadenum japonicum. J. Nat. Prod. 2015, 78, 258–264. [Google Scholar] [CrossRef]
- Delle Monache, F.; Delle Monache, G.; Pinheiro, R.M.; Radics, L. Chemistry of Clusia genus. Part 3. Nemorosonol, a derivative of tricyclo-[4.3.1.03,7]-decane-7-hydroxy-2,9-dione from Clusia nemorosa. Phytochemistry 1988, 27, 2305–2308. [Google Scholar] [CrossRef]
- Fan, Y.-M.; Yi, P.; Li, Y.; Yan, C.; Huang, T.; Gu, W.; Ma, Y.; Huang, L.J.; Zhang, J.X.; Yang, C.L.; et al. Two Unusual Polycyclic Polyprenylated Acylphloroglucinols, Including a Pair of Enantiomers from Garcinia multiflora. Org. Lett. 2015, 17, 2066–2069. [Google Scholar] [CrossRef]
- Tian, D.S.; Yi, P.; Xia, L.; Xiao, X.; Fan, Y.M.; Gu, W.; Huang, L.J.; Ben-David, Y.; Di, Y.T.; Yuan, C.M.; et al. Garmultins A-G, biogenetically related polycyclic Acylphloroglucinols from Garcinia multiflora. Org. Lett. 2016, 18, 5904–5907. [Google Scholar] [CrossRef]
- Li, Q.; Deng, A.J.; Qin, H.L.; Ji, M.; Li, Z.H.; Chen, X.G. Racemic 3,4-dihydro-4-naphthyl-naphthalen-1(2H)-ones from Juglans regia flowers. Fitoterapia 2019, 139, 104401. [Google Scholar] [CrossRef]
- Zhao, S.M.; Wang, Z.; Chen, X.Q.; Huang, M.B.; Tan, N.H. (±)-Rubioncolin D, a pair of enantiomeric naphthohydroquinone dimers from Rubia oncotricha. Tetrahedron Lett. 2017, 58, 3041–3043. [Google Scholar] [CrossRef]
- Zhao, S.M.; Wang, Z.; Zeng, G.Z.; Song, W.W.; Chen, X.Q.; Li, X.N.; Tan, N.H. New cytotoxic naphthohydroquinone dimers from Rubia alata. Org. Lett. 2014, 16, 5576–5579. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xie, L.Z.; Li, J.; Chen, G.D.; Aisa, H.A. A pair of new tetrahydro-naphthalenone enantiomers from Eremurus altaicus (Pall.). Stev. Phytochem. Lett. 2015, 13, 330–333. [Google Scholar] [CrossRef]
- Qiao, M.M.; Liu, F.; Liu, Y.; Guo, L.; Zhou, Q.M.; Peng, C.; Xiong, L. Curcumane C and (±)-curcumane D, an unusual seco-cadinane sesquiterpenoid and a pair of unusual nor-bisabolane enantiomers with significant vasorelaxant activity from Curcuma longa. Bioorg. Chem. 2019, 92, 103275. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zheng, R.R.; Liu, Z.W.; Wang, W.J.; Li, G.Q.; Fan, C.L.; Zhang, X.Q.; Ye, W.C.; Che, C.T. Enantiomeric chromones from Harrisonia perforata. Phytochem. Lett. 2014, 10, 295–299. [Google Scholar] [CrossRef]
- Yuan, W.J.; Gao, W.F.; Zhang, J.H.; Cao, P.; Zhang, Y.; Chen, D.Z.; Li, S.L.; Di, Y.T.; Hao, X.J. (±)-Perforison A, A Pair of New Chromone Enantiomers from Harrisonia perforata. Nat. Prod. Commun. 2017, 12, 63–65. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.F.; Han, C.; Xu, Q.-Q.; Wang, X.B.; Zhao, H.J.; Xue, G.M.; Luo, J.G.; Kong, L.Y. Isolation, chiral-phase resolution, and determination of the absolute configurations of a complete series of stereoisomers of a rearranged acetophenone with three stereocenters. J. Nat. Prod. 2019, 82, 1399–1404. [Google Scholar] [CrossRef]
- Tang, Y.Q.; Li, Y.Q.; Xie, Y.B.; Zhang, J.S.; Li, W.; Lou, L.L.; Zhang, G.; Yin, S. Evodialones A and B: Polyprenylated Acylcyclopentanone Racemates with a 3-Ethyl-1,1-diisopentyl-4-methylcyclopentane Skeleton from Evodia lepta. J. Nat. Prod. 2018, 81, 1483–1487. [Google Scholar] [CrossRef]
- Li, W.; Rao, L.; Liu, Y.; He, Q.; Fan, Y.; You, Y.-X.; Su, Y.; Hu, F.; Xu, Y.K.; Lin, B.; et al. (±)-Meliviticines A and B: Rearranged prenylated acetophenone derivatives from Melicope viticina and their antimicrobial activity. Bioorg. Chem. 2019, 90, 103099. [Google Scholar] [CrossRef]
- Wang, W.J.; Wang, L.; Huang, X.J.; Jiang, R.W.; Yang, X.L.; Zhang, D.M.; Chen, W.M.; Tang, B.Q.; Wang, Y.; Zhang, X.Q.; et al. Two pairs of new benzofuran enantiomers with unusual skeletons from Eupatorium chinense. Tetrahedron Lett. 2013, 54, 3321–3324. [Google Scholar] [CrossRef]
- Xu, J.F.; Zhao, H.J.; Wang, X.B.; Li, Z.R.; Luo, J.; Yang, M.H.; Yang, L.; Yu, W.Y.; Yao, H.Q.; Luo, J.G.; et al. (±)-Melicolones A and B, rearranged prenylated acetophenone stereoisomers with an unusual 9-oxatricyclo[3.2.1.13,8]nonane core from the leaves of Melicope ptelefolia. Org. Lett. 2015, 17, 146–149. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.J.; Zhu, L.J.; Zhao, Y.Q.; Zhen, Y.Q.; Zhang, L.; Lin, C.C.; Chen, L.X. Diarylheptanoid: A privileged structure in drug discovery. Fitoterapia 2020, 142. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, Z.L.; Huang, X.J.; Peng, Y.; Huang, X.; Shi, L.; Wang, Y.; Ye, W.C. Evaluation of diarylheptanoid-terpene adduct enantiomers from Alpinia officinarum for neuroprotective activities. J. Nat. Prod. 2018, 81, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.H.; Nikolic, D.; Simmler, C.; Qiu, F.; van Breemen, R.B.; Soejarto, D.D.; Pauli, G.F.; Chen, S.N. Diarylheptanoids from Dioscorea villosa (Wild Yam). J. Nat. Prod. 2012, 75, 2168–2177. [Google Scholar] [CrossRef] [Green Version]
- Mondal, S.; Verma, A.; Saha, S. Conformationally Restricted Triarylmethanes: Synthesis, Photophysical Studies, and Applications. Eur. J. Org. Chem. 2019, 2019, 864–894. [Google Scholar] [CrossRef]
- Zhou, B.; Yuan, X.J.; Li, J.Y.; Xu, Y.C.; Li, J.; Yue, J.M.; Liu, D.X.; Li, Y. (−)- and (+)-Securidanes A and B, Natural Triarylmethane Enantiomers: Structure and Bioinspired Total Synthesis. Research 2018, 2018, 2674182. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Yao, Y.; Huang, X.J.; Oberer, L.; Wagner, T.; Guo, J.M.; Gu, W.; Liu, W.D.; Lv, G.X.; Shen, Y.N.; et al. Four new selaginellin derivatives from Selaginella pulvinata: Mechanism of racemization process in selaginellins with quinone methide. Tetrahedron 2015, 71, 1581–1587. [Google Scholar] [CrossRef]
- Zou, J.; Chen, G.-D.; Zhao, H.; Huang, Y.; Luo, X.; Xu, W.; He, R.-R.; Hu, D.; Yao, X.S.; Gao, H. Triligustilides A and B: Two Pairs of Phthalide Trimers from Angelica sinensis with a Complex Polycyclic Skeleton and Their Activities. Org. Lett. 2018, 20, 884–887. [Google Scholar] [CrossRef]
- Mattio, L.M.; Catinella, G.; Pinto, A.; Dallavalle, S. Natural and nature-inspired stilbenoids as antiviral agents. Eur. J. Med. Chem. 2020, 202, 112541. [Google Scholar] [CrossRef]
- Li, X.L.; Zhao, B.X.; Huang, X.J.; Zhang, D.M.; Jiang, R.W.; Li, Y.J.; Jian, Y.Q.; Wang, Y.; Li, Y.L.; Ye, W.C. (+)- and (-)-Cajanusine, a Pair of New Enantiomeric Stilbene Dimers with a New Skeleton from the Leaves of Cajanus cajan. Org. Lett. 2014, 16, 224–227. [Google Scholar] [CrossRef]
- Ito, T.; Endo, H.; Shinohara, H.; Oyama, M.; Akao, Y.; Iinuma, M. Occurrence of stilbene oligomers in Cyperus rhizomes. Fitoterapia 2012, 83, 1420–1429. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Endo, H.; Oyama, M.; Iinuma, M. Novel isolation of stilbenoids with enantiomeric and meso forms from a cyperus rhizome. Phytochem. Lett. 2012, 5, 267–270. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, Z.M.; Yang, Y.N.; Jiang, J.S.; Zhang, P.C. Phenolic acid derivatives from Ligusticum chuanxiong. Phytochem. Lett. 2019, 33, 114–118. [Google Scholar] [CrossRef]
- Gao, Y.P.; Shen, Y.H.; Zhang, S.D.; Tian, J.M.; Zeng, H.W.; Ye, J.; Li, H.L.; Shan, L.; Zhang, W.-D. Incarvilleatone, a New Cyclohexylethanoid Dimer from Incarvillea younghusbandii and Its Inhibition against Nitric Oxide (NO) Release. Org. Lett. 2012, 14, 1954–1957. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.M.; Zheng, C.J.; Chen, G.Y.; Zhang, X.P.; Song, X.P.; Li, G.N.; Gan, L.S.; Sun, C.G. Bioactive Phenanthrene and Bibenzyl Derivatives from the Stems of Dendrobium nobile. J. Nat. Prod. 2016, 79, 1791–1797. [Google Scholar] [CrossRef]
- Malebo, H.M.; Kihampa, C.; Mgina, C.A.; Sung’hwa, F.; Jonker, S.A.; Nkunya, M.H.H.; Waibel, R. Antifungal Enantiomeric Styrylpyrones from Sanrafaelia ruffonammari and Ophrypetalum odoratum. Nat. Prod. Bioprospect. 2014, 4, 129–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, R.; Huang, X.; Zhang, Y.; Wu, L.; Nie, H.; Zhou, D.; Liu, B.; Deng, S.; Yang, R.; Huang, S.; et al. Structural Characterization and Assessment of the Cytotoxicity of 2,3-Dihydro-1H-indene Derivatives and Coumarin Glucosides from the Bark of Streblus indicus. J. Nat. Prod. 2016, 79, 2472–2478. [Google Scholar] [CrossRef]
- Wei, X.; Feng, C.; Li, X.H.; Mao, X.X.; Rong, L.; Yu, X.; Zhang, C.X.; Luo, H.B.; Zhang, D.M.; Ye, W.C.; et al. Enantiomeric Polyketides from the Starfish-Derived Symbiotic Fungus Penicillium sp. GGF16-1-2. Chem. Biodivers. 2019, 16, e1900052. [Google Scholar] [CrossRef]
- Ma, L.Y.; Zhang, H.B.; Kang, H.H.; Zhong, M.J.; Liu, D.S.; Liu, W.Z.; Ren, H. New Butenolides and Cyclopentenones from Saline Soil-Derived Fungus Aspergillus Sclerotiorum. Molecules 2019, 24, 2642. [Google Scholar] [CrossRef] [Green Version]
- He, J.W.; Wang, C.X.; Yang, L.; Chen, G.D.; Hu, D.; Guo, L.D.; Yao, X.S.; Gao, H. A Pair of New Polyketide Enantiomers from Three Endolichenic Fungal Strains Nigrospora sphaerica, Alternaria alternata, and Phialophora sp. Nat. Prod. Commun. 2016, 11, 829–831. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Fu, P.; Zhang, Y.; Liu, X.; Ren, F.; Che, Y. Sporulosol, a new ketal from the fungus Paraconiothyrium sporulosum. Molecules 2018, 23, 1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, J.; Li, X.M.; Li, X.; Li, H.L.; Meng, L.H.; Wang, B.-G. New lactone and isocoumarin derivatives from the marine mangrove-derived endophytic fungus Penicillium coffeae MA-314. Phytochem. Lett. 2019, 32, 1–5. [Google Scholar] [CrossRef]
- Liu, X.; Chen, C.; Zheng, Y.; Zhang, M.; Tong, Q.; Liu, J.; Zhou, Q.; Wang, J.; Luo, Z.; Zhu, H.; et al. (±)-Peniorthoesters A and B, Two Pairs of Novel Spiro-Orthoester en-antiomers With an Unusual 1,4,6-Trioxaspi-ro[4.5]decane-7-One Unit From Penicillium minioluteum. Front. Chem. 2018, 6, 605. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.J.; Chen, S.F.; Xu, X.W.; Zhao, D.; Wang, H.F.; Bai, J.; Hua, H.M.; Chen, G.; Pei, Y.H.; Lu, X.J.; et al. One pair of new cyclopentaisochromenone enantiomer from Alternaria sp. TNXY-P-1 and their cytotoxic activity. J. Asian Nat. Prod. Res. 2018, 20, 328–336. [Google Scholar] [CrossRef]
- Tang, J.W.; Xu, H.C.; Wang, W.G.; Hu, K.; Zhou, Y.F.; Chen, R.; Li, X.N.; Du, X.; Sun, H.D.; Puno, P.-T. (+)- And (-)-Alternarilactone A: Enantiomers with a Diepoxy-Cage-like Scaffold from an Endophytic Alternaria sp. J. Nat. Prod. 2019, 82, 735–740. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.B.; Li, T.X.; Yang, M.H.; Kong, L.Y. Bioactive metabolites from the endophytic fungus Alternaria alternata. Fitoterapia 2014, 99, 153–158. [Google Scholar] [CrossRef]
- Intaraudom, C.; Bunbamrung, N.; Dramae, A.; Boonyuen, N.; Choowong, W.; Rachtawee, P.; Pittayakhajonwut, P. Chromone derivatives, R- and S- taeniolin, from the marine-derived fungus Taeniolella sp. BCC31839. Nat. Prod. Res. 2021, 35, 392–398. [Google Scholar] [CrossRef]
- Wu, Y.H.; Xiao, G.K.; Chen, G.D.; Wang, C.X.; Hu, D.; Lian, Y.Y.; Lin, F.; Guo, L.D.; Yao, X.S.; Gao, H. Pericocins A-D, New Bioactive Compounds from Periconia sp. Nat. Prod. Commun. 2015, 10, 2127–2130. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.L.; Li, X.H.; Feng, D.; Jin, M.Y.; Cao, Y.M.; Cao, Z.X.; Deng, F.; Deng, Y.; Gu, Y.C.; Geng, Z. Novel Polyketides Produced by the Endophytic Fungus Aspergillus Fumigatus from Cordyceps Sinensis. Molecules 2018, 23, 1709. [Google Scholar] [CrossRef] [Green Version]
- Li, E.; Zhang, F.; Niu, S.; Liu, X.; Liu, G.; Che, Y. A Spiro[chroman-3,7’-isochromene]-4,6’(8’H)-dione from the Cordyceps-Colonizing Fungus Fimetariella sp. Org. Lett. 2012, 14, 3320–3323. [Google Scholar] [CrossRef]
- Li, X.H.; Han, X.H.; Qin, L.L.; He, J.L.; Cao, Z.X.; Guo, D.L.; Deng, Y.; Gu, Y.C. Isochromanes from Aspergillus fumigatus, an endophytic fungus from Cordyceps sinensis. Nat. Prod. Res. 2019, 33, 1870–1875. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Gao, W.; Zhang, M.; Li, Y.L.; Li, L.; Li, X.B.; Chang, W.Q.; Zhao, Z.T.; Lou, H.X. p-Terphenyl Derivatives from the Endolichenic Fungus Floricola striata. J. Nat. Prod. 2016, 79, 2188–2194. [Google Scholar] [CrossRef] [PubMed]
- Arunrattiyakorn, P.; Kuno, M.; Aree, T.; Laphookhieo, S.; Sriyatep, T.; Kanzaki, H.; Garcia Chavez, M.A.; Wang, Y.A.; Andersen, R.J. Biotransformation of β-Mangostin by an Endophytic Fungus of Garcinia mangostana to Furnish Xanthenes with an Unprecedented Heterocyclic Skeleton. J. Nat. Prod. 2018, 81, 2244–2250. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, S.; Qiu, P.; Tan, C.; Long, Y.; Lu, Y.; She, Z. (+)- and (-)-Ascomlactone A: A pair of novel dimeric polyketides from a mangrove endophytic fungus Ascomycota sp. SK2YWS-L. Org. Biomol. Chem. 2017, 15, 10276–10280. [Google Scholar] [CrossRef]
- Liu, Z.; Qiu, P.; Li, J.; Chen, G.; Chen, Y.; Liu, H.; She, Z. Anti-inflammatory polyketides from the mangrove-derived fungus Ascomycota sp. SK2YWS-L. Tetrahedron 2018, 74, 746–751. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, Z.; Lai, Y.; Li, D.; Wang, J.; Luo, Z.; Xue, Y.; Zhu, H.; Chen, C.; Zhang, Y. (±)-Terreinlactone A, a pair of 3-substituted δ-lactone enantiomers derived from terrein from the fungus Aspergillus terreus. Chem. Pharm. Bull. 2018, 66, 764–767. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.H.; Zhou, Y.H.; Xie, F.; Li, Y.L.; Zhao, Z.T.; Lou, H.X. Two new α-pyrone derivatives from an endolichenic fungus Tolypocladium sp. J. Asian. Nat. Prod. Res. 2017, 19, 786–792. [Google Scholar] [CrossRef]
- Sang, X.N.; Chen, S.F.; Chen, G.; An, X.; Li, S.G.; Lu, X.J.; Zhao, D.; Bai, J.; Wang, H.F.; Pei, Y.H. Two pairs of enantiomeric α-pyrone dimers from the endophytic fungus Phoma sp. YN02-P-3. RSC Adv. 2017, 7, 1943–1946. [Google Scholar] [CrossRef] [Green Version]
- Miyano, R.; Matsuo, H.; Nonaka, K.; Mokudai, T.; Niwano, Y.; Shiomi, K.; Takahashi, Y.; Omura, S.; Nakashima, T. Pochoniolides A and B, new antioxidants from the fungal strain Pochonia chlamydosporia var. spinulospora FKI-7537. J. Biosci. Bioeng. 2018, 126, 661–666. [Google Scholar]
- Qi, B.; Liu, X.; Mo, T.; Li, S.S.; Wang, J.; Shi, X.P.; Wang, X.H.; Zhu, Z.X.; Zhao, Y.F.; Jin, H.W.; et al. Nitric oxide inhibitory polyketides from Penicillium chrysogenum MT-12, an endophytic fungus isolated from Huperzia serrata. Fitoterapia 2017, 123, 35–43. [Google Scholar] [CrossRef]
- He, F.; Li, X.; Yu, J.H.; Zhang, X.; Nong, X.; Chen, G.; Zhu, K.; Wang, Y.Y.; Bao, J.; Zhang, H. Secondary metabolites from the mangrove sediment-derived fungus Penicillium pinophilum SCAU037. Fitoterapia 2019, 136, 104177. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.; Gong, Y.H.; Li, X.W.; Li, X.N.; Liu, J.J.; Chen, C.M.; Zhou, Y.; Gu, L.H.; Luo, Z.W.; Wang, J.P.; et al. Canescones A-E: Aromatic polyketide dimers with PTP1B inhibitory activity from Penicillium canescens. Org. Chem. Front. 2019, 6, 3274–3281. [Google Scholar] [CrossRef]
- Xu, L.L.; Chen, H.L.; Hai, P.; Gao, Y.; Xie, C.D.; Yang, X.L.; Abe, I. (+)- And (−)-Preuisolactone A: A Pair of Caged Norsesquiterpenoidal Enantiomers with a Tricyclo[4.4.01,6.02,8]decane Carbon Skeleton from the Endophytic Fungus Preussia isomera. Org. Lett. 2019, 21, 1078–1081. [Google Scholar] [CrossRef] [PubMed]
- Song, R.Y.; Liu, Y.; Liu, R.H.; Wang, X.B.; Li, T.X.; Kong, L.Y.; Yang, M.H. Benzophenone derivatives from the plant endophytic fungus, Pestalotiopsis sp. Phytochem. Lett. 2017, 22, 189–193. [Google Scholar] [CrossRef]
- Meng, L.H.; Li, X.M.; Liu, Y.; Wang, B.G.; Meng, L.H.; Liu, Y.; Mandi, A.; Kurtan, T. Isolation, Stereochemical Study, and Antioxidant Activity of Benzofuranone Derivatives from a Mangrove-derived Fungus Eurotium rubrum MA-150. Chirality 2016, 28, 581–584. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.Y.; Kuang, R.Q.; Chen, G.D.; Qin, S.Y.; Wang, C.X.; Hu, D.; Wu, B.; Liu, X.Z.; Yao, X.S.; Gao, H. Three pairs of new isopentenyl dibenzo[b,e]oxepinone enantiomers from Talaromyces flavus, a wetland soil-derived fungus. Molecules 2016, 21, 1184. [Google Scholar] [CrossRef] [Green Version]
- Hammerschmidt, L.; Ola, A.; Mueller, W.E.G.; Lin, W.; Mandi, A.; Kurtan, T.; Proksch, P.; Aly, A.H. Two new metabolites from the endophytic fungus Xylaria sp. isolated from the medicinal plant Curcuma xanthorrhiza. Tetrahedron Lett. 2015, 56, 1193–1197. [Google Scholar] [CrossRef]
- Liu, H.; Tan, H.; Wang, W.; Zhang, W.; Chen, Y.; Li, S.; Liu, Z.; Li, H.; Zhang, W. Cytorhizophins A and B, benzophenone-hemiterpene adducts from the endophytic fungus Cytospora rhizophorae. Org. Chem. Front. 2019, 6, 591–596. [Google Scholar] [CrossRef]
- Wu, J.C.; Hou, Y.; Xu, Q.; Fang, J.; Wu, Q.X.; Jin, X.J.; Chen, Y.; Hu, B. (±)-Alternamgin, a Pair of Enantiomeric Polyketides, from the Endophytic Fungi Alternaria sp. MG1. Org. Lett. 2019, 21, 1551–1554. [Google Scholar] [CrossRef]
- Zhong, W.M.; Wang, J.F.; Wei, X.Y.; Zeng, Q.; Chen, X.Y.; Xiang, Y.; Tian, X.P.; Zhang, S.; Long, L.J.; Wang, F.-Z. (+)- And (−)-Eurotone A: A pair of enantiomeric polyketide dimers from a marine-derived fungus Eurotium sp. SCSIO F452. Tetrahedron Lett. 2019, 60, 1600–1603. [Google Scholar] [CrossRef]
- Shaker, S.; Fan, R.Z.; Lan, W.J.; Li, H.J. A pair of novel bisindole alkaloid enantiomers from marine fungus Fusarium sp. XBB-9. Nat. Prod. Res. 2021, 35, 1497–1503. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, L.; Wang, B.; Xu, Y.; Zhu, G.; Lan, M.; Zhu, W.; Sun, K. Diketopiperazine and diphenylether derivatives from marine algae-derived Aspergillus versicolor OUCMDZ-2738 by epigenetic activation. Mar. Drugs 2019, 17, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, J.; Li, X.M.; Meng, L.H.; Konuklugil, B.; Li, X.; Li, H.L.; Wang, B.G. Isolation and characterization of three pairs of indolediketopiperazine enantiomers containing infrequent N-methoxy substitution from the marine algal-derived endophytic fungus Acrostalagmus luteoalbus TK-43. Bioorg. Chem. 2019, 90, 103030. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Wang, J.; Wei, X.; Fu, T.; Chen, Y.; Zeng, Q.; Huang, Z.; Huang, X.; Zhang, W.; Zhang, S.; et al. Three pairs of new spirocyclic alkaloid enantiomers from the marine-derived fungus Eurotium sp. SCSIO F452. Front. Chem. 2019, 7, 350. [Google Scholar] [CrossRef]
- Gao, H.; Liu, W.; Zhu, T.; Mo, X.; Mandi, A.; Kurtan, T.; Li, J.; Ai, J.; Gu, Q.; Li, D. Diketopiperazine alkaloids from a mangrove rhizosphere soil derived fungus Aspergillus effuses H1-1. Org. Biomol. Chem. 2012, 10, 9501–9506. [Google Scholar] [CrossRef] [Green Version]
- Cai, R.; Jiang, H.; Xiao, Z.; Liu, Z.; Lin, S.e.; She, Z.; Cao, W.; Yan, T.; Long, Y.; She, Z. (−)- and (+)-Asperginulin A, a Pair of Indole Diketopiperazine Alkaloid Dimers with a 6/5/4/5/6 Pentacyclic Skeleton from the Mangrove Endophytic Fungus Aspergillus sp. SK-28. Org. Lett. 2019, 21, 9633–9636. [Google Scholar] [CrossRef]
- Chen, G.D.; Bao, Y.R.; Huang, Y.F.; Hu, D.; Li, X.X.; Guo, L.D.; Li, J.; Yao, X.S.; Gao, H. Three pairs of variecolortide enantiomers from Eurotium sp. with caspase-3 inhibitory activity. Fitoterapia 2014, 92, 252–259. [Google Scholar] [CrossRef]
- Zhong, W.; Wang, J.; Wei, X.; Chen, Y.; Fu, T.; Xiang, Y.; Huang, X.; Tian, X.; Xiao, Z.; Zhang, W.; et al. Variecolortins A-C, Three Pairs of Spirocyclic Diketopiperazine Enantiomers from the Marine-Derived Fungus Eurotium sp. SCSIO F452. Org. Lett. 2018, 20, 4593–4596. [Google Scholar] [CrossRef]
- Han, J.; Liu, C.; Li, L.; Zhou, H.; Liu, L.; Bao, L.; Chen, Q.; Song, F.; Zhang, L.; Li, E.; et al. Decalin-Containing Tetramic Acids and 4-Hydroxy-2-pyridones with Antimicrobial and Cytotoxic Activity from the Fungus Coniochaeta cephalothecoides Collected in Tibetan Plateau (Medog). J. Org. Chem. 2017, 82, 11474–11486. [Google Scholar] [CrossRef]
- Li, J.; Wang, W.X.; Chen, H.P.; Li, Z.H.; He, J.; Zheng, Y.S.; Sun, H.; Huang, R.; Yuan, Q.X.; Feng, T.; et al. (±)-Xylaridines A and B, Highly Conjugated Alkaloids from the Fungus Xylaria longipes. Org. Lett. 2019, 21, 1511–1514. [Google Scholar] [CrossRef]
- Li, J.; Wang, W.X.; Li, Z.H.; He, J.; Huang, R.; Zheng, Y.S.; Li, L.Q.; Wang, X.; Feng, T.; Liu, J.K. Xylaridines C and D, unusual thiopyranodipyridine alkaloids from the fungus Xylaria longipes. Org. Lett. 2019, 21, 6145–6148. [Google Scholar] [CrossRef]
- Chen, S.; Jiang, M.; Chen, B.; Niaz, S.I.; He, J.; Liu, L.; Chen, S.; Liu, L.; Salaenoi, J.; Niaz, S.I. Penicamide A, A Unique N,N’-Ketal Quinazolinone Alkaloid from Ascidian-Derived Fungus Penicillium sp. 4829. Mar. Drugs 2019, 17, 522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Li, F.; Ji, N. Alkaloids from an algicolous strain of Talaromyces sp. Chin. J. Oceanol. Limnol. 2016, 34, 367–371. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.K.; Gao, T.; Yang, M.Y.; Zhao, G.Z.; Zhu, H.J.; Cao, F.; Zhang, B.; Liu, L. A pair of enantiomeric 5-oxabicyclic[4.3.0]lactam derivatives and one new polyketide from the marine-derived fungus Penicillium griseofulvum. Nat. Prod. Res. 2018, 32, 2366–2369. [Google Scholar] [CrossRef] [PubMed]
- Almeida, C.; Hemberger, Y.; Schmitt, S.M.; Bouhired, S.; Natesan, L.; Kehraus, S.; Dimas, K.; Gutschow, M.; Bringmann, G.; Konig, G.M. Marilines A-C: Novel phthalimidines from the sponge-derived fungus Stachylidium sp. Chemistry 2012, 18, 8827–8834. [Google Scholar] [CrossRef]
- Kong, Z.; Jing, R.; Geng, Y.; Ji, J.; Wu, Y.; Guo, Y.; Qin, L.; Zheng, C. Trichodermadiones A and B from the solid culture of Trichoderma atroviride S361, an endophytic fungus in Cephalotaxus fortunei. Fitoterapia 2018, 127, 362–366. [Google Scholar] [CrossRef]
- Wen, H.; Li, Y.; Liu, X.; Ye, W.; Yao, X.; Che, Y. Fusagerins A-F, New Alkaloids from the Fungus Fusarium sp. Nat. Prod. Bioprospect. 2015, 5, 195–203. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Chen, X.; Li, D.; Zhang, Y.; Li, L.; Guo, L.; Cao, Y.; Che, Y. Bisabolane Sesquiterpenoids from the Plant Endophytic Fungus Paraconiothyrium brasiliense. J. Nat. Prod. 2015, 78, 746–753. [Google Scholar] [CrossRef]
- Jia, Y.L.; Wei, M.Y.; Guan, F.F.; Wang, C.Y.; Shao, C.L.; Jia, Y.L.; Wei, M.Y.; Chen, H.Y. (+)- and (−)-Pestaloxazine A, a Pair of Antiviral Enantiomeric Alkaloid Dimers with a Symmetric Spiro[oxazinane-piperazinedione] Skeleton from Pestalotiopsis sp. Org. Lett. 2015, 17, 4216–4219. [Google Scholar] [CrossRef]
- Yan, Y.M.; Zhang, H.X.; Liu, H.; Wu, J.B.; Li, Y.P.; Cheng, Y.X.; Wang, Y. (+/−)-Lucidumone, a COX-2 Inhibitory Caged Fungal Meroterpenoid from Ganoderma lucidum. Org. Lett. 2019, 21, 8523–8527. [Google Scholar] [CrossRef]
- Peng, X.R.; Liu, J.Q.; Wan, L.S.; Li, X.N.; Yan, Y.X.; Qiu, M.H. Four New Polycyclic Meroterpenoids from Ganoderma cochlear. Org. Lett. 2014, 16, 4838–4841. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.M.; Ai, J.; Zhou, L.L.; Chung, A.C.K.; Li, R.; Nie, J.; Fang, P.; Wang, X.L.; Luo, J.; Hu, Q.; et al. Lingzhiols, Unprecedented Rotary Door-Shaped Meroterpenoids as Potent and Selective Inhibitors of p-Smad3 from Ganoderma lucidum. Org. Lett. 2013, 15, 5488–5491. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, L.; Li, S.; Zhao, J. Meroterpenoids from the fruiting bodies of higher fungus Ganoderma resinaceum. Phytochem. Lett. 2017, 22, 214–218. [Google Scholar] [CrossRef]
- Luo, Q.; Di, L.; Yang, X.H.; Cheng, Y.X. Applanatumols A and B, meroterpenoids with unprecedented skeletons from Ganoderma applanatum. RSC Adv. 2016, 6, 45963–45967. [Google Scholar] [CrossRef]
- Luo, Q.; Wang, X.L.; Di, L.; Yan, Y.M.; Lu, Q.; Yang, X.H.; Hu, D.B.; Cheng, Y.X. Isolation and identification of renoprotective substances from the mushroom Ganoderma lucidum. Tetrahedron 2015, 71, 840–845. [Google Scholar] [CrossRef]
- Wang, X.F.; Yan, Y.M.; Wang, X.L.; Ma, X.J.; Fu, X.Y.; Cheng, Y.X. Two new compounds from Ganoderma lucidum. J. Asian Nat. Prod. Res. 2015, 17, 329–332. [Google Scholar] [CrossRef]
- Luo, Q.; Tu, Z.C.; Cheng, Y.X. Two rare meroterpenoidal rotamers from Ganoderma applanatum. RSC Adv. 2017, 7, 3413–3418. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Wang, F.; Xu, F.; Ding, L.Q.; Zhang, Q.; Li, H.X.; Zhao, F.; Wang, L.Q.; Zhu, L.H.; Chen, L.X.; et al. Two pairs of farnesyl phenolic enantiomers as natural nitric oxide inhibitors from Ganoderma sinense. Bioorg. Med. Chem. Lett. 2016, 26, 3342–3345. [Google Scholar] [CrossRef]
- Li, L.; Li, H.; Peng, X.R.; Hou, B.; Yu, M.Y.; Dong, J.R.; Li, X.N.; Zhou, L.; Yang, J.; Qiu, M.H. (±)-Ganoapplanin, a Pair of Polycyclic Meroterpenoid Enantiomers from Ganoderma applanatum. Org. Lett. 2016, 18, 6078–6081. [Google Scholar] [CrossRef]
- Luo, Q.; Wang, Z.; Luo, J.-F.; Tu, Z.C.; Cheng, Y.X. (±)-Applanatumines B-D: Novel dimeric meroterpenoids from Ganoderma applanatum as inhibitors of JAK3. RSC Adv. 2017, 7, 38037–38043. [Google Scholar] [CrossRef] [Green Version]
- Qin, F.Y.; Cheng, Y.X.; Qin, F.Y.; Cheng, Y.X.; Yan, Y.M.; Cheng, Y.X.; Tu, Z.C. (±) Gancochlearols A and B: Cytotoxic and COX-2 inhibitory meroterpenoids from Ganoderma cochlear. Nat. Prod. Res. 2020, 34, 2269–2275. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.Y.; Cheng, Y.X.; Qin, F.Y.; Cheng, Y.X.; Yan, Y.M.; Cheng, Y.X.; Tu, Z.C. (±) Cochlearoids N-P: Three pairs of phenolic meroterpenoids from the fungus Ganoderma cochlear and their bioactivities. J. Asian Nat. Prod. Res. 2019, 21, 542–550. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.J.; Nian, Y.; Yan, Y.; Gong, Y.; Luo, Q.; Zhang, Y.; Hou, B.; Zuo, Z.L.; Wang, S.M.; Jiang, H.H.; et al. Two new classes of T-type calcium channel inhibitors with new chemical scaffolds from Ganoderma cochlear. Org. Lett. 2015, 17, 3082–3085. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Liu, F.; Su, H.G.; Guo, L.; Zhou, Q.M.; Huang, Y.J.; Peng, C.; Xiong, L. Two pairs of alkaloid enantiomers from Ganoderma luteomarginatum. Biochem. Syst. Ecol. 2019, 86, 103930. [Google Scholar] [CrossRef]
- Nord, C.; Menkis, A.; Broberg, A. Cytotoxic illudane sesquiterpenes from the fungus Granulobasidium vellereum (Ellis and Cragin) Jülich. J. Nat. Prod. 2015, 78, 2559–2564. [Google Scholar] [CrossRef]
- McMorris, T.C.; Kelner, M.J.; Chadha, R.K.; Siegel, J.S.; Moon, S.S.; Moya, M.M. Structure and reactivity of illudins. Tetrahedron 1989, 45, 5433–5440. [Google Scholar] [CrossRef]
- Kinder, F.R., Jr.; Bair, K.W. Total Synthesis of (±)-Illudin M. J. Org. Chem. 1994, 59, 6965–6967. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, L.; Fu, X.; Li, Z.; Guo, L.; Kou, L.; Liu, M.; Xie, Z. (+)- and (−)-actinoxocine, and actinaphthorans A-B, C-ring expansion and cleavage angucyclinones from a marine-derived Streptomyces sp. Org. Chem. Front. 2019, 6, 3925–3928. [Google Scholar] [CrossRef]
- Yi, W.; Li, Q.; Song, T.; Chen, L.; Li, X.C.; Zhang, Z.; Lian, X.Y. Isolation, structure elucidation, and antibacterial evaluation of the metabolites produced by the marine-sourced Streptomyces sp. ZZ820. Tetrahedron 2019, 75, 1186–1193. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, Q.; Guo, L.; Zhang, Y.; Feng, L.; Zhou, L.; Yang, S.; Yao, Q.; Pescitelli, G.; Xie, Z. Isolation, structure elucidation and racemization of (+)- and (−)-pratensilins A-C: Unprecedented spiro indolinone-naphthofuran alkaloids from a marine Streptomyces sp. Chem. Commun. 2017, 53, 10066–10069. [Google Scholar] [CrossRef]
- Liang, Y.Q.; Liao, X.J.; Zhao, B.X.; Xu, S.H.; Liang, Y.Q.; Xu, S.H. (+)- and (−)-Spongiterpene, a pair of new valerenane sesquiterpene enantiomers from the marine sponge Spongia sp. Nat. Prod. Res. 2021, 35, 2178–2183. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.Y.; Han, G.Y.; Yang, N.N.; Lan, L.F.; Li, X.W.; Guo, Y.W. Racemic trinorsesquiterpenoids from the Beihai sponge Spongia officinalis: Structure and biomimetic total synthesis. Org. Chem. Front. 2018, 5, 1022–1027. [Google Scholar] [CrossRef]
- Jiao, W.H.; Hong, L.L.; Sun, J.B.; Piao, S.J.; Chen, G.D.; Deng, H.; Wang, S.P.; Yang, F.; Lin, H.-W. (±)-Hippolide J, A Pair of Unusual Antifungal Enantiomeric Sesterterpenoids from the Marine Sponge Hippospongia lachne. Eur. J. Org. Chem. 2017, 2017, 3421–3426. [Google Scholar] [CrossRef]
- Afifi, A.H.; Kagiyama, I.; El-Desoky, A.H.; Kato, H.; Mangindaan, R.E.P.; de Voogd, N.J.; Ammar, N.M.; Hifnawy, M.S.; Tsukamoto, S. Sulawesins A-C, Furanosesterterpene Tetronic Acids That Inhibit USP7, from a Psammocinia sp. Marine Sponge. J. Nat. Prod. 2017, 80, 2045–2050. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, Y.; Gu, B.B.; Yang, F.; Jiao, W.H.; Hu, G.H.; Yu, H.B.; Han, B.N.; Zhang, W.; Shen, Y.; et al. Antifungal bromopyrrole alkaloids from the South China Sea sponge Agelas sp. Tetrahedron 2016, 72, 2964–2971. [Google Scholar] [CrossRef]
- Allen, E.E.; Zhu, C.; Panek, J.S.; Schaus, S.E. Multicomponent Condensation Reactions via ortho-Quinone Methides. Org. Lett. 2017, 19, 1878–1881. [Google Scholar] [CrossRef] [Green Version]
- Chu, M.J.; Tang, X.L.; Qin, G.F.; Sun, Y.T.; Li, L.; de Voogd, N.J.; Li, P.L.; Li, G.Q. Pyrrole Derivatives and Diterpene Alkaloids from the South China Sea Sponge Agelas nakamurai. Chem. Biodivers. 2017, 14. [Google Scholar] [CrossRef]
- Wang, Q.; Tang, X.; Luo, X.; de Voogd, N.J.; Li, P.; Li, G. (+)- and (−)-Spiroreticulatine, A Pair of Unusual Spiro Bisheterocyclic Quinoline-imidazole Alkaloids from the South China Sea Sponge Fascaplysinopsis reticulata. Org. Lett. 2015, 17, 3458–3461. [Google Scholar] [CrossRef]
- Shirouzu, T.; Watari, K.; Ono, M.; Koizumi, K.; Saiki, I.; Tanaka, C.; van Soest, R.W.M.; Miyamoto, T. Structure, synthesis, and biological activity of a C-20 bisacetylenic alcohol from a marine sponge Callyspongia sp. J. Nat. Prod. 2013, 76, 1337–1342. [Google Scholar] [CrossRef]
- Jang, K.H.; Lee, Y.; Sim, C.J.; Oh, K.B.; Shin, J. Bioactive lipids from the sponge Spirastrella abata. Bioorg. Med. Chem. Lett. 2012, 22, 1078–1081. [Google Scholar] [CrossRef]
- Alam, N.; Wang, W.; Hong, J.; Lee, C.-O.; Im, K.S.; Jung, J.H. Cytotoxic sphingosine 4-sulfates from the sponge Spirastrella abata. J. Nat. Prod. 2002, 65, 944–945. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.M.; Li, L.J.; Qin, X.C.; Lu, Q.; Tu, Z.C.; Cheng, Y.X. Compounds from the insect Blaps japanensis with COX-1 and COX-2 inhibitory activities. Bioorg. Med. Chem. Lett. 2015, 25, 2469–2472. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.M.; Ai, J.; Shi, Y.N.; Zuo, Z.L.; Hou, B.; Luo, J.; Cheng, Y.X. (±)-Aspongamide A, an N-Acetyldopamine Trimer Isolated from the Insect Aspongopus chinensis, Is an Inhibitor of p-Smad3. Org. Lett. 2014, 16, 532–535. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.J.; Yan, Y.M.; Tu, Z.C.; Luo, J.F.; Liang, R.; Yang, T.H.; Cheng, Y.X.; Wang, S.M. Compounds from Polyphaga plancyi and their inhibitory activities against JAK3 and DDR1 kinases. Fitoterapia 2016, 114, 163–167. [Google Scholar] [CrossRef]
- Chan, S.T.S.; Nani, R.R.; Schauer, E.A.; Martin, G.E.; Williamson, R.T.; Sauri, J.; Buevich, A.V.; Schafer, W.A.; Joyce, L.A.; Goey, A.K.L.; et al. Characterization and Synthesis of Eudistidine C, a Bioactive Marine Alkaloid with an Intriguing Molecular Scaffold. J. Org. Chem. 2016, 81, 10631–10640. [Google Scholar] [CrossRef]
- Wang, L.; Ma, Y.T.; Sun, Q.Y.; Li, X.N.; Yan, Y.; Yang, J.; Yang, F.M.; Liu, F.Y.; Zang, Z.; Wu, X.H.; et al. Structurally diversified diterpenoids from Euphorbia dracunculoides. Tetrahedron 2015, 71, 5484–5493. [Google Scholar] [CrossRef]
- Marin-Saez, J.; Romero-Gonzalez, R.; Garrido, F.A. Enantiomeric determination and evaluation of the racemization process of atropine in Solanaceae seeds and contaminated samples by high performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2016, 1474, 79–84. [Google Scholar] [CrossRef] [Green Version]
- Capon, R.J. Extracting value: Mechanistic insights into the formation of natural product artifacts-case studies in marine natural products. Nat. Prod. Rep. 2020, 37, 55–79. [Google Scholar] [CrossRef]
- Li, L.-Z.; Sun, X.; Qi, X.-L.; Song, S.-J.; Liang, X.; Wang, J.; Zhao, Q.C.; Song, J. Bioactive norditerpenoids and neolignans from the roots of salvia miltiorrhiza. Org. Biomo.l Chem. 2016, 14, 10050–10057. [Google Scholar] [CrossRef]
- Song, X.-Q.; Zhu, K.; Yu, J.-H.; Zhang, Q.; Zhang, Y.; He, F.; Cheng, Z.-Q.; Jiang, C.-S.; Bao, J.; Zhang, H. New Octadecanoid Enantiomers from the Whole Plants of Plantago depressa. Molecules 2018, 23, 1723. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Jiang, J.; Liu, Z.; Lin, S.; Xia, G.; Xia, X.; Ding, B.; He, L.; Lu, Y.; She, Z. Peniphenones A-D from the mangrove fungus Penicillium dipodomyicola HN4-3A as inhibitors of Mycobacterium tuberculosis phosphatase MptpB. J. Nat. Prod. 2014, 77, 800–806. [Google Scholar] [CrossRef]
- Cao, W.-W.; Luo, Q.; Cheng, Y.-X.; Wang, S.-M. Meroterpenoid enantiomers from Ganoderma sinensis. Fitoterapia 2016, 110, 110–115. [Google Scholar] [CrossRef]
- Zhang, X.; Li, P.-L.; Qin, G.-F.; Li, S.; de Voogd, N.J.; Tang, X.-L.; Li, G.Q. Isolation and absolute configurations of diversiform C17, C21 and C25 terpenoids from the marine sponge Cacospongia sp. Mar. Drugs 2019, 17, 14. [Google Scholar] [CrossRef] [Green Version]


















































| No. | C-7 & C-8 Configurations | J7,8 Values | Specific Optical Rotations | ECD Data | ||||
|---|---|---|---|---|---|---|---|---|
| J7,8 (Hz) | Solvent | [α]D | Solvent | T (°C) | Δε | λ (nm) | ||
| 1a | (7R,8S)-erythro | 3.5 | CD3OD | +3.3 | MeOH | 25 | +2.25 | 239 |
| 1b | (7S,8R)-erythro | 3.5 | CD3OD | −4.7 | MeOH | 25 | −2.75 | 239 |
| 2a | (7R,8R)-threo | 6.8 | CD3OD | −21.7 | MeOH | 25 | −3.45 | 231 |
| 2b | (7S,8S)-threo | 6.8 | CD3OD | +16.0 | MeOH | 25 | +2.83 | 232 |
| 3a | (7S,8S)-threo | 8.8 | CDCl3 | +18.0 | MeOH | 20 | −1.12 | 239 |
| 3b | (7R,8R)-threo | 8.8 | CDCl3 | −20.0 | MeOH | 20 | +1.40 | 237 |
| 4a | (7R,8S)-erythro | 2.6 | CDCl3 | +20.5 | MeOH | 20 | +2.38 | 245 |
| 4b | (7S,8R)-erythro | 2.6 | CDCl3 | −22.0 | MeOH | 20 | −1.89 | 243 |
| 5a | (7R,8S)-erythro | 3.3 | CDCl3 | +32.0 | MeOH | 20 | +0.25 | 238 |
| 5b | (7S,8R)-erythro | 3.3 | CDCl3 | −28.2 | MeOH | 20 | −0.01 | 238 |
| 6a | (7S,8S)-threo | 6.9 | CDCl3 | +36.7 | MeOH | 20 | +8.92 | 239 |
| 6b | (7R,8R)-threo | 6.9 | CDCl3 | −33.5 | MeOH | 20 | −6.50 | 240 |
| 7a | (7R,8R)-threo | 8.0 | CDCl3 | −28.5 | MeOH | 20 | +9.64 | 230 |
| 7b | (7S,8S)-threo | 8.0 | CDCl3 | +26.9 | MeOH | 20 | −8.74 | 230 |
| 8a | (7R,8S)-erythro | 7.2 | CDCl3 | −31.0 | MeOH | 20 | −3.55 | 240 |
| 8b | (7S,8R)-erythro | 7.2 | CDCl3 | +29.0 | MeOH | 20 | +2.80 | 240 |
| 9a | (7S,8S)-threo | 7.4 | CDCl3 | +34.0 | MeOH | 20 | +3.70 | 244 |
| 9b | (7R,8R)-threo | 7.4 | CDCl3 | −34.0 | MeOH | 20 | −2.81 | 243 |
| 10a | (7R,8R)-threo | 6.5 | CDCl3 | −28.0 | MeOH | 20 | +16.90 | 240 |
| 10b | (7S,8S)-threo | 6.5 | CDCl3 | +32.0 | MeOH | 20 | −17.65 | 238 |
| 11a | (7R,8R)-threo | 7.6 | CDCl3 | −20.0 | MeOH | 20 | −6.57 | 238 |
| 11b | (7S,8S)-threo | 7.6 | CDCl3 | +21.0 | MeOH | 20 | +8.87 | 242 |
| 12a | (7R,8S)-erythro | 4.7 | CDCl3 | −31.0 | MeOH | 20 | −3.55 | 240 |
| 12b | (7S,8R)-erythro | 4.7 | CDCl3 | +29.0 | MeOH | 20 | +2.80 | 240 |
| 13a | (7S,8R)-erythro | 3.0 | CDCl3 | +17.1 | CHCl3 | 20 | −1.03 | 232 |
| 13b | (7R,8S)-erythro | 3.0 | CDCl3 | −16.2 | CHCl3 | 20 | +1.03 | 232 |
| 14a | (7S,8S)-threo | 6.3 | CDCl3 | −35.1 | CHCl3 | 20 | −1.14 | 240 |
| 14b | (7R,8R)-threo | 6.3 | CDCl3 | +32.6 | CHCl3 | 20 | +1.17 | 240 |
| 15a | (7R,8R)-threo | 8.1 | CDCl3 | +30.4 | CHCl3 | 20 | +1.58 | 240 |
| 15b | (7S,8S)-threo | 8.1 | CDCl3 | −29.8 | CHCl3 | 20 | −1.54 | 240 |
| 16a | (7S,8R)-erythro | 3.3 | CDCl3 | +16.6 | CHCl3 | 20 | +3.18 | 234 |
| 16b | (7R,8S)-erythro | 3.3 | CDCl3 | −16.4 | CHCl3 | 20 | −3.20 | 234 |
| 17a | (7R,8R)-threo | 8.0 | CDCl3 | +7.0 | MeOH | 20 | - | - |
| 17b | (7S,8S)-threo | 8.0 | CDCl3 | −7.0 | MeOH | 20 | - | - |
| 18a | (7S,8R)-erythro | 4.7 | CDCl3 | +20.0 | MeOH | 20 | +11.17 | 230 |
| 18b | (7R,8S)-erythro | 4.7 | CDCl3 | −18.0 | MeOH | 20 | −4.00 | 235 |
| 19a | (7S,8S)-threo | 6.1 | CD3OD | +18.0 | MeOH | 20 | +1.70 | 230 |
| 19b | (7R,8R)-threo | 6.1 | CD3OD | −18.0 | MeOH | 20 | −1.20 | 230 |
| 20a | (7R,8S)-erythro | 4.8 | CD3OD | −10.0 | MeOH | 20 | +2.70 | 232 |
| 20b | (7S,8R)-erythro | 4.8 | CD3OD | +8.0 | MeOH | 20 | −2.82 | 233 |
| 21a | (7S,8R)-erythro | 5.4 | CD3OD | +15.0 | MeOH | 20 | +3.15 | 228 |
| 21b | (7R,8S)-erythro | 5.4 | CD3OD | −15.0 | MeOH | 20 | −2.65 | 228 |
| 22a | (7S,8S)-threo | 7.7 | CDCl3 | +17.2 | MeOH | 20 | −0.59 | 233 |
| 22b | (7R,8R)-threo | 7.7 | CDCl3 | −19.0 | MeOH | 20 | +0.51 | 232 |
| 23a | (7R,8R)-threo | 7.6 | CDCl3 | −24.5 | MeOH | 20 | - | - |
| 23b | (7S,8S)-threo | 7.6 | CDCl3 | +26.0 | MeOH | 20 | - | - |
| 24a | (7R,8S)-erythro | 4.6 | CDCl3 | −18.0 | MeOH | 20 | +12.20 | 241 |
| 24b | (7S,8R)-erythro | 4.6 | CDCl3 | +22.0 | MeOH | 20 | −10.81 | 241 |
| 25a | (7S,8S)-threo | 6.8 | CDCl3 | +23.0 | MeOH | 20 | +0.57 | 240 |
| 25b | (7R,8R)-threo | 6.8 | CDCl3 | −25.0 | MeOH | 20 | −1.05 | 237 |
| Cell lines | Compds. | IC50 (μM) | Reference | Cell Lines | Compds. | IC50 (μM) | Reference |
|---|---|---|---|---|---|---|---|
| (a) | |||||||
| HL-60 | (+)-171a | 5.62 | [105] | HCT-116 | (−)-314a | 1.38 | [162] |
| (−)-171b | 3.51 | [105] | (+)-314b | >10 | [162] | ||
| (±)-171 | 2.65 | [105] | HepG2 | (−)-314a | 3.30 | [162] | |
| (+)-172a | 9.64 | [105] | (+)-314b | >10 | [162] | ||
| (−)-172b | 8.16 | [105] | BGC-823 | (−)-314a | 6.51 | [162] | |
| (±)-172 | 5.58 | [105] | (+)-314b | >10 | [162] | ||
| Hep3B | (−)-268a | >10 | [146] | NIC-H1650 | (−)-314a | 8.19 | [162] |
| (+)-268b | 5.1 | [146] | (+)-314b | >10 | [162] | ||
| HL-60 | (+)-297a | 12.08 | [156] | A2780 | (−)-314a | 2.14 | [162] |
| (−)-297b | 19.24 | [156] | (+)-314b | >10 | [162] | ||
| MDA-MB-231 | (+)-297a | >50 | [156] | SF-268 | (+)-536a | 12.5 | [261] |
| (−)-297b | 18.46 | [156] | (−)-536b | >100 | [261] | ||
| HCT-116 | (+)-312a | inactive | [160] | (+)-537a | 30.1 | [261] | |
| (−)-312b | 14.23 | [160] | (−)-537b | >100 | [261] | ||
| A549 | (−)-390a | 4.64 | [8] | HepG2 | (+)-536a | 15.0 | [261] |
| (+)-390b | 10.54 | [8] | (−)-536b | >100 | [261] | ||
| MCF-7 | (−)-390a | 5.60 | [8] | (+)-537a | 37.3 | [261] | |
| (+)-390b | 15.52 | [8] | (−)-537b | >100 | [261] | ||
| MDA-MB-231 | (−)-390a | 3.86 | [8] | ||||
| (+)-390b | 11.86 | [8] | |||||
| (b) | |||||||
| HL-60 | (+)-381a | 3.42 | [191,192] | MCF-7 | (+)-381a | 4.18 | [191,192] |
| (−)-381b | >20 | [191,192] | (−)-381b | >20 | [191,192] | ||
| (−)-382a | 16.54 | [191,192] | (−)-382a | 14.44 | [191,192] | ||
| (+)-382b | >40 | [191,192] | (+)-382b | >40 | [191,192] | ||
| (±)-382 | 14.44 | [191,192] | (±)-382 | 36.00 | [191,192] | ||
| (−)-383a | 3.15 | [191,192] | (−)-383a | 5.85 | [191,192] | ||
| (+)-383b | 2.35 | [191,192] | (+)-383b | 10.76 | [191,192] | ||
| (±)-383 | 18.08 | [191,192] | (±)-383 | 17.05 | [191,192] | ||
| (−)-384a | 3.45 | [191,192] | (−)-384a | 3.17 | [191,192] | ||
| (+)-384b | 2.36 | [191,192] | (+)-384b | 3.08 | [191,192] | ||
| (±)-384 | 2.93 | [191,192] | (±)-384 | 11.92 | [191,192] | ||
| (−)-385a | 2.63 | [191,192] | (−)-385a | 14.60 | [191,192] | ||
| (+)-385b | 5.41 | [191,192] | (+)-385b | 15.02 | [191,192] | ||
| (±)-385 | 13.90 | [191,192] | (±)-385 | 15.47 | [191,192] | ||
| SMMC-7721 | (+)-381a | 4.19 | [191,192] | SW480 | (+)-381a | 7.22 | [191,192] |
| (−)-381b | >20 | [191,192] | (−)-381b | >20 | [191,192] | ||
| (−)-382a | 16.20 | [191,192] | (−)-382a | 17.43 | [191,192] | ||
| (+)-382b | >40 | [191,192] | (+)-382b | >40 | [191,192] | ||
| (±)-382 | 22.83 | [191,192] | (±)-382 | 27.73 | [191,192] | ||
| (−)-383a | 5.55 | [191,192] | (−)-383a | 3.04 | [191,192] | ||
| (+)-383b | 12.30 | [191,192] | (+)-383b | 5.53 | [191,192] | ||
| (±)-383 | 20.71 | [191,192] | (±)-383 | 13.60 | [191,192] | ||
| (−)-384a | 3.80 | [191,192] | (−)-384a | 1.99 | [191,192] | ||
| (+)-384b | 10.83 | [191,192] | (+)-384b | 1.52 | [191,192] | ||
| (±)-384 | 15.36 | [191,192] | (±)-384 | 7.62 | [191,192] | ||
| (−)-385a | 12.53 | [191,192] | (−)-385a | 3.39 | [191,192] | ||
| (+)-385b | 13.21 | [191,192] | (+)-385b | 4.31 | [191,192] | ||
| (±)-385 | 14.82 | [191,192] | (±)-385 | 8.84 | [191,192] | ||
| A549 | (+)-381a | 4.51 | [191,192] | A549 | (±)-383 | 24.02 | [191,192] |
| (−)-381b | >20 | [191,192] | (−)-384a | 2.96 | [191,192] | ||
| (−)-382a | 17.27 | [191,192] | (+)-384b | 18.78 | [191,192] | ||
| (+)-382b | >40 | [191,192] | (±)-384 | 15.44 | [191,192] | ||
| (±)-382 | 17.30 | [191,192] | (−)-385a | 22.36 | [191,192] | ||
| (−)-383a | 4.40 | [191,192] | (+)-385b | 9.53 | [191,192] | ||
| (+)-383b | 16.33 | [191,192] | (±)-385 | 10.24 | [191,192] | ||
| Virus/Host | Compds. | EC50 (μM) | IC50 (μM) | SI | Reference |
|---|---|---|---|---|---|
| HSV-1/Vero | (+)-254a | 6.41 | 19.25 | 3.0 | [7] |
| (−)-254b | 3.70 | 16.02 | 4.3 | [7] | |
| (±)-254 | 1.23 | 2.14 | 1.7 | [7] | |
| Acyclovir b | >100 | 0.41 | >243.9 | [7] | |
| Virus A (H3N3)/MDCK | (+)-104a | 9.86 | 19.25 | 1.9 | [67] |
| (−)-104b | 11.11 | 77.61 | 6.9 | [67] | |
| (±)-104 | 3.13 | 6.42 | 2.1 | [67] | |
| (−)-105a | 8.62 | 77.61 | 7.9 | [67] | |
| (+)-105b | 17.46 | 57.74 | 3.3 | [67] | |
| (±)-105 | 2.87 | 25.87 | 9.0 | [67] | |
| Oseltamivir b | 3.38 | 3073 | 910.5 | [67] | |
| Ribavirin b | 6.19 | 4771 | 770.7 | [67] | |
| KSHV/Vero | (+)-368a | 8.75 | 140.6 | 16.06 | [184] |
| (−)-368b | 29.13 | 173.7 | 5.96 | [184] | |
| (+)-369a | 202.9 | >500 | >2.46 | [184] | |
| (−)-369b | 140.9 17.67 | >500 | >2.55 | [184] | |
| (+)-370a | 211.1 | 12.51 | [184] | ||
| (−)-370b | 39.80 | >300 | >7.50 | [184] | |
| (+)-371a | 40.00 | >300 | >7.50 | [184] | |
| (−)-371b | 158.50 | >300 | >1.89 | [184] | |
| Acyclovir b | 0.41 | 99.18 | 241.9 | [184] | |
| EV71/Vero | (−)-558a | 69.1 | 143.7 | 2.1 | [272] |
| (+)-558b | 14.2 | 130.2 | 9.2 | [272] | |
| (±)-558 | 14.2 | 126.6 | 7.9 | [272] | |
| Ribavirin b | >256.1 | 4098 | >16 | [272] |
| Bacterial Strains | Compds. | Activities | Reference |
|---|---|---|---|
| Enterococcus faecalis | (+)-147a | MIC = 21.97 μg/mL | [97] |
| (−)-147b | MIC = 12.54 μg/mL | [97] | |
| Penicilin a | MIC < 2.96 μg/mL | [97] | |
| Staphylococcus aureus | (+)-285a | IC50 = 1.27 μM | [153] |
| (−)-285b | IC50 = 1.79 μM | [153] | |
| (+)-286a | IC50 = 2.27 μM | [153] | |
| (−)-286b | IC50 = 4.3 μM | [153] | |
| (+)-287a | IC50 = 3.6 μM | [153] | |
| (−)-287b | IC50 = 6.0 μM | [153] | |
| (+)-288a | IC50 = 0.61 μM | [153] | |
| (−)-288b | IC50 = 2.27 μM | [153] | |
| Chloramphenicol a | IC50 = 0.43 μM | [153] |
| Fungal Strains | Compds. | Activities | Reference |
|---|---|---|---|
| Candida albicans | (−)-464a | MIC = 26.2 μM | [219] |
| (+)-464b | inactive | [219] | |
| Candida albicans | (+)-483a | MIC80 = 19.5 μg/mL | [229] |
| (−)-483b | MIC80 = 48.8 μg/mL | [229] | |
| (+)-484a | MIC80 = 24.0 μg/mL | [229] | |
| (−)-484b | MIC80 > 50.0 μg/mL | [229] | |
| Fusarium solani | (+)-524a | MIC > 64 μg/mL | [256] |
| (−)-524b | MIC = 32 μg/mL | [256] |
| Assay Model | Compds. | IC50 (μM) | Reference |
|---|---|---|---|
| BV-2/NO | (+)-43a | 26.4 | [39] |
| (−)-43b | 8.9 | [39] | |
| (+)-44a | 5.9 | [39] | |
| (−)-44b | 14.7 | [39] | |
| Quercetin a | 17.0 | [39] | |
| RAW 264.7/NO | (+)-82a | 17.9 | [54] |
| (−)-82b | 5.6 | [54] | |
| (+)-83a | 15.1 | [54] | |
| (−)-83b | 4.3 | [54] | |
| RAW 264.7/NO | (+)-221a | 3.6 | [124] |
| (−)-221b | 22.8 | [124] | |
| (±)-221 | 14.7 | [124] | |
| Indomethacin a | 42.2 | [124] | |
| BV-2/NO | (+)-407a | 6.9 | [130] |
| (−)-407b | 1.4 | [130] | |
| (±)-407 | 1.0 | [130] | |
| Minocyclinebe a | 27.2 | [130] |
| Assay Model | Compds. | IC50 (μM) | Reference |
|---|---|---|---|
| DPPH | (+)-527a | 5.8 | [257] |
| (−)-527b | 23.5 | [257] | |
| (+)-528a | 9.8 | [257] | |
| (−)-528b | 24.9 | [257] | |
| (+)-529a | 3.7 | [257] | |
| (−)-529b | 6.1 | [257] | |
| Ascorbic acid a | 23.0 | [257] |
| Cell/Inducing Agents | Compds. | Cell Viability | Reference |
|---|---|---|---|
| SH-SY5Y/H2O2 | (−)-24a | 54.7% at 50 μM | [21] |
| (+)-24b | 70.5% at 50 μM | [21] | |
| Trolox a | ~69.0% at 50 μM | [21] | |
| SH-SY5Y/H2O2 | (+)-72a | ~69% at 25 μM | [46] |
| (−)-72b | inactive at 25 μM | [46] | |
| Trolox a | ~62% at 25 μM | [46] | |
| SH-SY5Y/H2O2 | (+)-119a | 76.29% at 50 μM | [75] |
| (−)-119b | 56.48% at 50 μM | [75] | |
| PC12/OKA | (+)-129a | 65.4% at 10 μM | [83] |
| (−)-129b | 83.4% at 10 μM | [83] | |
| (+)-130a | 91.2% at 10 μM | [83] | |
| (−)-130b | 69.5% at 10 μM | [83] | |
| H9C2/ischemia-hypoxia | (−)-165a | ~51% at 0.1 μM | [103] |
| (+)-165b | inactive | [103] | |
| (−)-166a | ~55% at 0.1 μM | [103] | |
| (+)-166b | ~45% at 0.1 μM | [103] | |
| (−)-173a | inactive | [103] | |
| (+)-173b | ~52% at 0.1 μM | [103] | |
| Salvianolic acid B a | ~46% at 0.1 μM | [103] | |
| HUVEC/glucose | (+)-423a | 102.6% at 1 μM | [200] |
| (−)-423b | 79.9% at 1 μM | [200] | |
| (+)-426a | 102.6% at 1 μM | [200] | |
| (−)-426b | 79.9% at 1 μM | [200] | |
| Cortical neurons/MPP+ | (+)-433a | ~90% at 16 μM | [206] |
| (−)-433b | inactive | [206] |
| Enzymes | Compds. | IC50 (μM) | Reference |
|---|---|---|---|
| AchE | (+)-157a | >100 | [100] |
| (−)-157b | 28.3 | [100] | |
| Galanthamine a | 1.9 | [100] | |
| AchE | (+)-523a | 2.3 | [256] |
| (−)-523b | 13.8 | [256] | |
| (±)-523 | 9.5 | [256] | |
| Tacrine a | 0.14 | [256] | |
| α-Glucosidase | (+)-495a | 63.7 | [237] |
| (−)-495b | 27.9 | [237] | |
| (±)-495 | 36.1 | [237] | |
| Acarbose a | 477.0 | [237] | |
| PTP1B | (−)-339a | Inactive | [176] |
| (+)-339b | 43.6 | [176] | |
| (+)-340a | 38.1 | [176] | |
| (−)-340b | Inactive | [176] | |
| (−)-341a | 61.0 | [176] | |
| (+)-341b | Inactive | [176] | |
| (−)-342a | 58.2 | [176] | |
| (+)-342b | Inactive | [176] | |
| Oleanolic acid a | 2.5 | [176] | |
| COX-2 | (+)-641a | 2.52 | [305] |
| (−)-641b | 6.04 | [305] | |
| (+)-644a | 17.8 | [305] | |
| (−)-644b | 9.7 | [305] | |
| Celecoxib a | 0.016 | [305] |
| Compds. | Inhibition (%) | Reference |
|---|---|---|
| (+)-6a | 31.2 | [19] |
| (−)-6b | 75.3 | [19] |
| Curcumin a | 62.1 | [19] |
| (+)-73a | 62.1 | [51] |
| (−)-73b | 81.6 | [51] |
| Curcumin a | 63.2 | [51] |
| (+)-132a | 85.8 | [85] |
| (−)-132b | 73.6 | [85] |
| Curcumin a | 57.0 | [85] |
| (+)-242a | 33.9 | [131] |
| (−)-242b | 50.6 | [131] |
| Curcumin a | 63.3 | [131] |
| Models | Compds. | Activities | Reference |
|---|---|---|---|
| MT1 receptor agonistic activity | (−)-145a | agonistic rate = 11.26% | [93] |
| (+)-145b | inactive | [93] | |
| MT2 receptor agonistic activity | (−)-145a | agonistic rate = 52.44% | [93] |
| (+)-145b | inactive | [93] | |
| NF-κB inhibition | (+)-330a | inactive | [169] |
| (−)-330b | IC50 = 7.27 μM | [169] | |
| Antifouling activity | (−)-530a | inactive | [259] |
| (+)-530b | adhesive rate = 48.4% | [259] | |
| Protein-protein interaction inhibition | (+)-647a | inactive | [308] |
| (−)-647b | modestly | [308] | |
| Olfactory responses | (+)-648a | strong | [309] |
| (−)-648b | weak | [309] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.-H.; Yu, Z.-P.; Capon, R.J.; Zhang, H. Natural Enantiomers: Occurrence, Biogenesis and Biological Properties. Molecules 2022, 27, 1279. https://doi.org/10.3390/molecules27041279
Yu J-H, Yu Z-P, Capon RJ, Zhang H. Natural Enantiomers: Occurrence, Biogenesis and Biological Properties. Molecules. 2022; 27(4):1279. https://doi.org/10.3390/molecules27041279
Chicago/Turabian StyleYu, Jin-Hai, Zhi-Pu Yu, Robert J. Capon, and Hua Zhang. 2022. "Natural Enantiomers: Occurrence, Biogenesis and Biological Properties" Molecules 27, no. 4: 1279. https://doi.org/10.3390/molecules27041279