- 1Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan, China
- 2Joint Center for Single Cell Biology, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
- 3Institute of Cash Crops, Hubei Academy of Agricultural Sciences, Wuhan, China
- 4Xinjiang Key Laboratory of Special Species Conservation and Regulatory Biology, College of Life Science, Xinjiang Normal University, Ürümqi, China
Multiple C2 domain and transmembrane region proteins (MCTPs) are a group of evolutionarily conserved proteins and show emerging roles in mediating protein trafficking and signaling transduction. Although, several studies showed that MCTPs play important roles during plant growth and development, their biological functions in cotton remain largely unknown. Here, we identify and characterize 33 GhMCTP genes from upland cotton (Gossypium hirsutum) and reveal the diverse expression patterns of GhMCTPs in various tissues. We also find that GhMCTP7, GhMCTP12, and GhMCTP17 are highly expressed in the main stem apex, suggesting their possible roles in shoot development. Through analyzing different cotton species, we discover plant heights are closely related to the expression levels of GhMCTP7, GhMCTP12, and GhMCTP17. Furthermore, we silence the expression of GhMCTP genes using virus-induced gene silencing (VIGS) system in cotton and find that GhMCTP7, GhMCTP12, and GhMCTP17 play an essential role in shoot meristem development. GhMCTPs interact with GhKNAT1 and GhKNAT2 and regulate meristem development through integrating multiple signal pathways. Taken together, our results demonstrate functional redundancy of GhMCTPs in cotton shoot meristem development and provide a valuable resource to further study various functions of GhMCTPs in plant growth and development.
Introduction
The development of multicellular organisms is regulated by various signaling pathways. These signaling transduction events are mediated by extensive intercellular and intracellular membrane trafficking processes, which control the signal perception and the trafficking of signal molecules from one compartment to another. Thus, membrane trafficking regulates the processing, modification, and secretion of signal molecules, or conversely, their translocation to the nucleus (Shilo and Schejter, 2011; Van Norman et al., 2011).
C2 domain is one of the most prevalent eukaryotic lipid-binding domains and could serve as a docking module that targets proteins to a specific intracellular membrane (Nalefski and Falke, 1996; Cho and Stahelin, 2006). A large number of C2 domain-containing proteins have been identified, and most of them are involved in membrane trafficking and signal transduction (Corbalan-Garcia and Gómez-Fernández, 2014). Multiple C2 domain and transmembrane region proteins (MCTPs) are evolutionarily conserved in eukaryotic organisms, containing 3–4 C2 domains at the N-terminus and transmembrane regions at the C-terminus (Shin et al., 2005; Lek et al., 2012). MCTPs mediate the trafficking of key regulators and are essential for signaling transduction in diverse species, thus regulating various developmental processes (Liu et al., 2013; Genç et al., 2017). MCTPs also function as unique membrane tethers controlling endoplasmic reticulum (ER)-plasma membrane (PM) contact specifically at plasmodesmata and regulate cell-to-cell communication (Brault et al., 2019).
The function of MCTP was first identified in Caenorhabditis elegans in a high-throughput RNAi screening. Genetic mutants of MCTP were embryonic lethal (Maeda et al., 2001). In addition, different alleles of mctp mutants in Drosophila were isolated and showed to regulate various developmental processes, including larval development and neurotransmission, suggesting multiple roles of MCTPs in different developmental stages (Tunstall et al., 2012; Genç et al., 2017). However, the molecular functions of MCTPs in regulating these processes were still largely unknown.
The invertebrate organisms C. elegans and Drosophila melanogaster contain a single MCTP gene. In all plant lineages, the number of MCTP repertoire significantly increases, and each of MCTPs exhibits distinct or overlapping patterns of gene expression and subcellular protein localization (Liu et al., 2018a; Hao et al., 2020; Zhu et al., 2020), suggesting more diverse and specific functions of MCTPs in regulating multiple cellular and developmental processes. Several members of MCTP proteins have been identified in plants to mediate the intercellular and intracellular trafficking of various macromolecules. QUIRKY (QKY) and FT-INTERACTING PROTEIN 1 (FTIP1) belong to the MCTP family, mediate the trafficking of florigen protein FLOWERING LOCUS T (FT) from companion cells to sieve elements, thus regulating flowering time in Arabidopsis (Liu et al., 2012, 2019). QKY also interacts with and stabilizes a leucine-rich repeat receptor-like kinase SCRAMBLED (SCM), and is required for proper cell-type patterning and organogenesis (Trehin et al., 2013; Vaddepalli et al., 2014; Song et al., 2019; Mergner et al., 2020). In addition, two other MCTP proteins, FTIP3 and FTIP4, interact with key meristem regulator SHOOTMERISTEMLESS (STM) and control its subcellular localization and intercellular trafficking in the shoot apex, thus determining the fate of shoot apical meristem (Liu et al., 2018b).
Multiple C2 domain and transmembrane region proteins also regulate multiple developmental processes in other plant species. OsFTIP1, the closet ortholog of FTIP1 in rice, mediates the flowering transition by affecting the trafficking of RICE FLOWERING LOCUS T1 (RFT1) from companion cells to sieve elements (Song et al., 2017). OsFTIP1 also determines the nuclear localization of rice MOTHER OF FT AND TFL1 (OsMFT1) and promotes drought tolerance (Chen et al., 2021). Another MCTP protein in rice OsFTIP7 facilitates nuclear translocation of a homeodomain transcription factor, Oryza sativa homeobox 1 (OSH1), to determine the auxin-mediated anther dehiscence in rice (Song et al., 2018). ZmCpd33, the closet homolog of QKY in maize, promotes sucrose export from companion cells into sieve elements (Tran et al., 2019). DOFTIP1, the orchid orthologs of FTIP1, plays an important role in promoting flowering in the orchid Dendrobium Chao Praya Smile (Wang et al., 2017). These results demonstrate that MCTPs are involved in diverse protein trafficking events and regulate plant development.
Upland cotton (Gossypium hirsutum) is an important economic crop in the world, and it is the primary fiber crop and an important oil crop (John and Crow, 1992). The architecture of cotton plants is determined primarily by their plant heights, shoot branching patterns, and flowering patterns, all of which directly affect cotton planting strategies, yield, planting area, mechanized harvesting suitability, and cotton planting costs (Reinhardt and Kuhlemeier, 2002; Su et al., 2018). In G. hirsutum, GhMCTPs have been genome-widely identified and the gene expression patterns have been analyzed (Hao et al., 2020); however, the biological functions of GhMCTPs in G. hirsutum are still largely unknown.
In this study, we systematically investigated MCTPs in cotton and analyzed their gene expressions in various developing cotton tissues. We further characterized the function of GhMCTP7, GhMCTP12, and GhMCTP17 in shoot development. GhMCTP7, GhMCTP12, and GhMCTP17 show distinct or overlapping subcellular localization patterns in Nicotiana benthamiana leaf epidermal cells. They interact with GhKNAT1 and GhKNAT2 and regulate shoot apical meristem development through integrating multiple signaling pathways. Our results demonstrate the functional redundancy of GhMCTPs in shoot development and reveal a gene regulatory framework that determines the meristem fate, providing a valuable resource for cotton architecture improvement.
Materials and Methods
Plant Materials
The seeds of upland cotton (G. hirsutum “coker 312”) were surface sterilized with 70% (v/v) ethanol for 1min, and then with 10% hydrogen peroxide for 2h, followed by washing with sterile water several times. The sterilized seeds were germinated on one-half strength Murashige and Skoog (MS) medium (12-h-light/12-h-dark cycle, 28°C), and seedlings were transplanted to the soil for further growth. The roots, stems, main stem apex (MSA, apex length is about 5mm), young leaves of three-leaf stage cotton plants, and 10DPA (days post-anthesis) cotton fiber after flowering were harvested for RNA extraction.
The shoot apexes (about 1cm) of eight upland cotton cultivars (G. hirsutun Okra, Emian JD1718, Lumian 1, Jimian 958, Emian JB2150, Emian SJA146, Emian JC1751, Zaosong2) were harvested for RNA extraction when these plants were flowering. The height of plants was calculated for each cultivar (n=50) when cotton bolls were open.
Sequence Analysis
A BLASTP search was performed using the protein sequences of 16 Arabidopsis MCTPs as query sequences on the website of COTTONGEN1 by chosen the G. hirsutum (AD1) ZJUv2.1 proteins (totally 72,761 protein sequences). The proteins with high sequence similarity (E-value=0) were selected as putative GhMCTPs. All GhMCTP sequences were then manually searched against MotifScan,2 InterProScan,3 and SMART,4 to confirm the sequence containing the C2 domain and PRT_C domain. Finally, a total of 33 GhMCTP members were identified. The chromosomal location, amino acid length, protein molecular mass, and isoelectric point of the 33 GhMCTPs were analyzed using COTTONGEN5 and ExPASy ProtParam.6 DNA and protein sequences were analyzed using DNASTAR software (DNAStar, MD, United States).
Phylogenetic Analysis
The protein sequences of 33 GhMCTPs were used as queries to identify GhMCTP homologs in Gossypium raimondii and Gossypium arboretum from COTTONGEN,7 and different plant species from Phytozome v12.8 A phylogenetic tree of deduced GhMCTP amino acid sequences was constructed using the neighbor-joining algorithm with default parameters, with 1,000 bootstrap replicates in MEGA_X_10.2.4 (Kumar et al., 2016).9
Gene Structure and Chromosomal Mapping
The Gene Structure Display Server Program10 was used to draw the exon-intron structure of GhMCTP genes based on the full-length genome sequence and the corresponding coding sequences. Domain analysis of all MCTP proteins from various plant species was performed by InterProScan. The chromosomal location information of all MCTP genes was derived from the annotation information downloaded on the COTTONGEN (see footnote 5; Supplementary Table 1).11 Based on the location information of GhMCTPs, we manually drew the chromosome map using Photoshop software.
Heat-Map Analysis of Gene Expression
The reads per kb per million reads (RPKM) values denoting the expression levels of GhMCTP genes were obtained from a comprehensive profile of the TM-1 transcriptome data (Trapnell et al., 2012; Zhang et al., 2015),12 and the expression data of main stem apex were generated in this study. A heat-map analysis was performed using TBtools (Chen et al., 2020).13
Expression Analysis
Total RNA was extracted from cotton roots, stems, leaves, ovules, and the 10days fiber after flowering using the RNAprep Pure Plant kit (TIANGEN, Beijing, China) and reverse transcribed using Moloney Murine Leukemia Virus Reverse Transcriptase (Promega, Madison, Wisconsin, United States) according to the manufacturer’s instructions. Quantitative real-time PCR was performed using MJ Research DNA Engine Option 2 detection system with the fluorescent intercalating dye SYBR-Green (Toyobo). The relative expression levels were normalized to a cotton polyubiquitin gene (GhUBI1, GenBank accession no. EU604080).
A two-step PCR procedure was performed in all experiments using the previously described method (Huang et al., 2013). The expressions of the putative target genes were determined using the comparative cycle threshold method. To achieve optimal amplification, PCR conditions for each set of primers were optimized for annealing temperature and Mg2+ concentration. Data presented in the quantitative real-time PCR (qRT-PCR) analysis are the mean and SD of three biological replicates of plant materials and three technical replicates in each biological sample using gene-specific primers. Primers used for qRT-PCR are designed to target GhMCTP-A and GhMCTP-D simultaneously and are listed in Supplementary Table 2.
Yeast Two-Hybrid Assay
The N-terminal fragments of GhMCTP7, GhMCTP12, and GhMCTP17 devoid of the sequences encoding the transmembrane regions were amplified and cloned into pGADT7 (Prey vector, Clontech). The coding sequences of GhKNAT1 and GhKNAT2 were amplified and cloned into pGBKT7 (Bait vector, Clontech). The prey and bait vectors were transformed into AH109 and Y187 cells, respectively. After mating, all transformed cells were grown on a Synthetic Defined-Ade/-His/-Trp/-Leu medium for interaction tests. Primers used are listed in Supplementary Table 2.
Luciferase Complementation Imaging Assay
The coding sequences of GhMCTP7, GhMCTP12, and GhMCTP17 were amplified and cloned into the N-terminal Luciferase fusion vector JW771-N-terminal luciferase fragment (nLUC). The coding sequences of GhKNAT1 and GhKNAT2 were amplified and cloned into the C-terminal Luciferase fusion vector JW772-C-terminal luciferase fragment (cLUC). The resulting plasmids were transformed into Agrobacterium cells, which were infiltrated into N. benthamiana leaves. The luciferase complementation imaging (LCI) assay was performed as described previously (Chen et al., 2008). The LUC luminescence signal was visualized using a cryogenically cooled CCD camera (Night Shade LB985, Berthold Technologies) and indiGO software. Primers used are listed in Supplementary Table 2.
Agrobacterium tumefaciens-Mediated VIGS
Overnight Agrobacterium cultures with the desired tobacco rattle virus (TRV) vectors, including TRV2:GhMCTP7, TRV2:GhMCTP12, TRV2:GhMCTP17, and TRV2:GhKNAT1/2, were infiltrated into two fully expanded cotyledons of 10-day-old cotton plants grown at 22–24°C as described previously (Gao et al., 2013). At least 15 plants were inoculated for each construct. The TRV2:GhCLA1 construct was included as a visual marker for virus-induced gene silencing (VIGS) efficiency. To simultaneously silence the expression of GhMCTP7, GhMCTP12, and GhMCTP17, Agrobacterium cultures containing TRV2:GhMCTP7, TRV2:GhMCTP12, and TRV2:GhMCTP17 were mixed and infiltrated. Primers used are listed in Supplementary Table 2.
Results
Identification of MCTP Genes in Cotton
As MCTPs were first identified in Arabidopsis in plants (Liu et al., 2018a), we used 16 MCTP protein sequences in Arabidopsis as queries to search against the protein database of G. hirsutum to identify putative GhMCTPs in cotton using the BLASTP program in COTTONGEN [G. hirsutum (AD1) ZJUv2.1 proteins (72761)]. The identified MCTP members were further used as queries to search for other possible MCTP proteins in G. hirsutum. All these putative GhMCTP proteins were subjected for domain analysis to confirm that all these candidates contain 3–4 N-terminal C2 domains and phosphoribosyltransferase C-terminal region (PRT_C). Finally, we identified a total of 33 GhMCTP proteins in G. hirsutum (Figure 1).
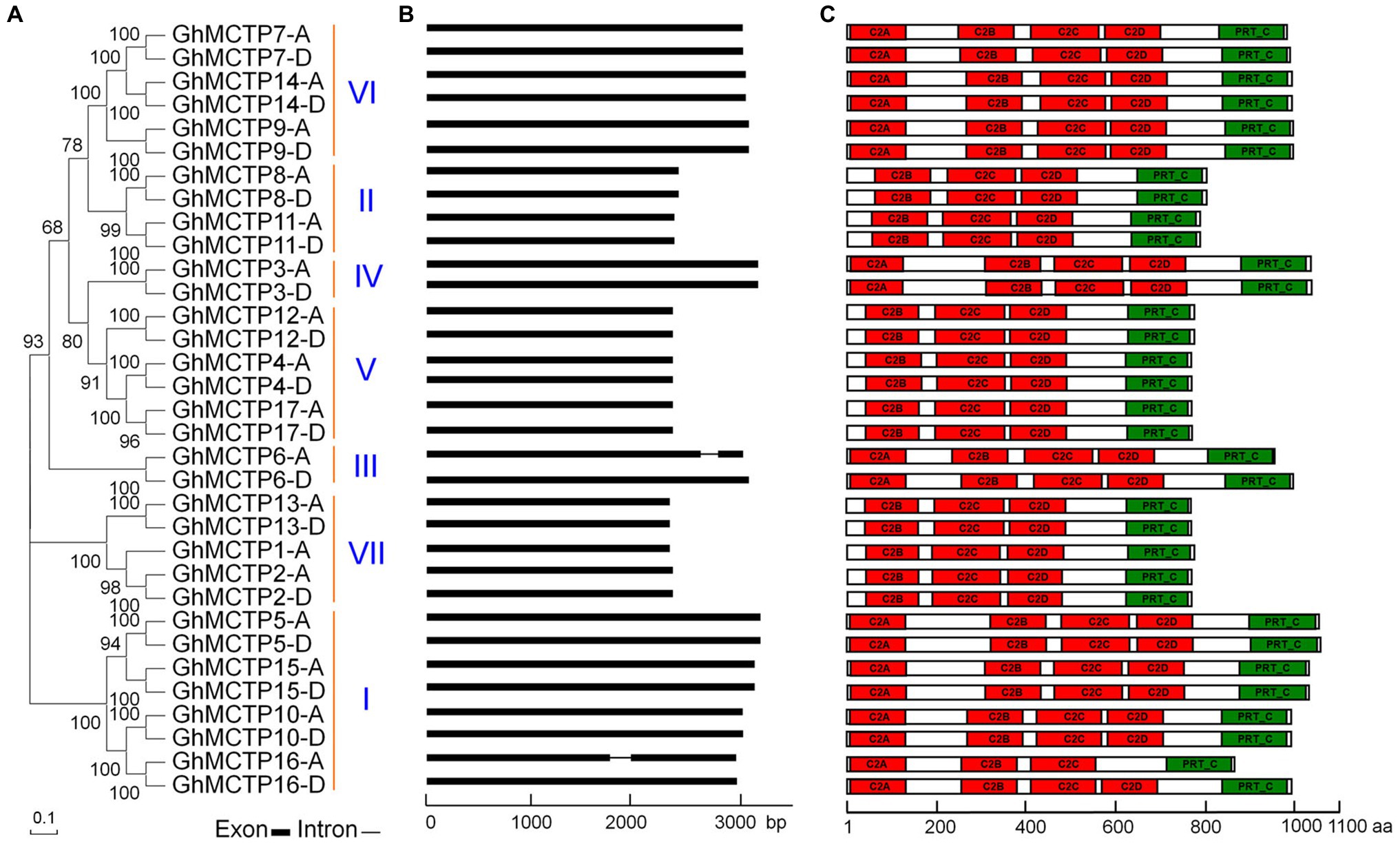
Figure 1. Characterization of multiple C2 domain and transmembrane region proteins (MCTP) family proteins in upland cotton (Gossypium hirsutum). (A) Thirty-three GhMCTP proteins are classified into seven groups based on phylogenetic analysis of MCTP proteins in G. hirsutum. The phylogenetic tree was generated with MEGA X using the neighbor-joining algorithm. Numbers on the major branches indicate bootstrap values (>50%) in 1,000 replicates. (B) Schematic diagrams showing the gene structures of GhMCTP genes. The coding regions are indicated by black boxes. The intron is represented by a black line. (C) Protein motif analysis of GhMCTPs. The prediction of protein motifs is based on InterProScan. C2 domain and Phosphoribosyltransferase C-terminal are labeled as red and green boxes, respectively. bp, base pair; aa, amino acids.
We classified these 33 GhMCTPs into seven clades through phylogenetic analysis based on multiple sequence alignment (Figure 1A). In addition, we aligned all these GhMCTP protein sequences and identified 16 pairs of MCTP protein with high sequence similarity. The members from each pair are originated from the cotton A subgenome (At, where ‘t’ stands from tetraploid) and D subgenome (Dt), respectively. The GhMCTPs were therefore classified as GhMCTP-A and GhMCTP-D. The members in these two subgroups were further designated according to their locations on the chromosome (Supplementary Figure 1; Supplementary Table 1), according to the naming principle in other cotton genome studies (Paterson et al., 2012; Huang et al., 2020).
We analyzed the gene structures of all GhMCTPs and found most of GhMCTP genes do not contain introns (Figure 1B). Similar to MCTPs in Arabidopsis, most GhMCTP proteins contain 1–4 C-terminal transmembrane regions except for GhMCTP3 (Supplementary Figure 2), suggesting the functional conservation and divergence among MCTPs. We also searched the MCTP proteins in the diploid cotton Gossypium arboreum and Gossypium raimondii (Paterson et al., 2012; Huang et al., 2020), and identified 17 and 18 MCTP members from G. arboreum genome (A-genome) and G. raimondii genome (D-genome), respectively (Supplementary Table 1).
Phylogenetic Analysis of MCTP Homologs in Plants
To identify MCTP homologs in plant genomes, we performed BLASTP or TBLASTN search in protein and genome databases in Phytozome 12 using MCTP1 as a query sequence. We obtained MCTP homologs in different land plants and algae including dicotyledons (G. hirsutum, G. arboreum, G. raimondii, Arabidopsis, Aquilegia coerulea, Amaranthus hypochondriacus, olanum lycopersicum, Eucalyptus grandis, Populus trichocarpa, and Medicago truncatula), monocotyledons (Ananas comosus, Oryza sativa, and Zea mays), moss (Physcomitrella patens), fern (Selaginella moellendorffii), and algaes (Chlamydomonas reinhardtii and Micromonas pusilla; Supplementary Table 3). To understand the evolutionary relationships among MCTPs in plants, MCTP homologs in different species were analyzed in detail using neighbor-joining and maximum likelihood methods, and the unrooted phylogenetic tree was constructed (Figure 2A).
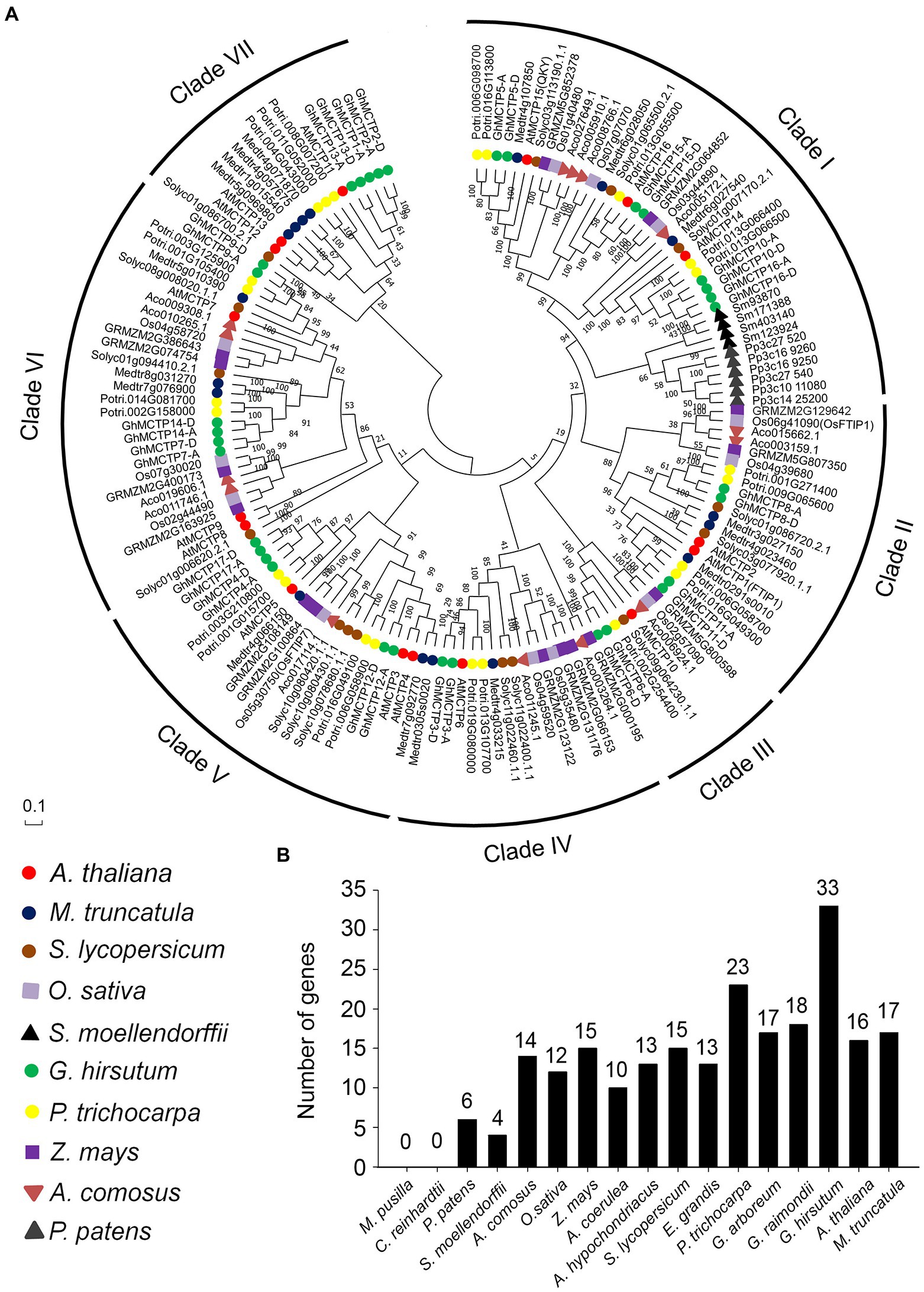
Figure 2. Polygenetic relationships of MCTP homologs in different plant species. (A) Proteins from 10 different species (Arabidopsis thaliana, Medicago truncatula, Solanum lycopersicum, Oryza sativa, Selaginella moellendorffii, G. hirsutum, Populus trichocarpa, Zea mays, Ananas comosus, and Physcomitrella patens) are indicated by different icons and are classified into seven groups. All available gene names are also indicated. The level of statistical support was conducted by neighbor-joining method, and numbers on the major branches indicate bootstrap values. (B) Numbers of MCTP genes in different species.
The MCTP homologs could be classified into seven clades, namely clade I to clade VII. Each clade contains MCTPs from eudicot plants (Figure 2A). The clade I was the largest branch containing 41 members, which are from different plant species including eudicots, monocots, fern, and moss. Clade III is the smallest one with only eight members. Furthermore, clade VII is specific for eudicots, and clade I is the only clade containing MCTPs from moss (P. patens) and fern (S. moellendorffii; Figure 2A). It is noteworthy that MCTPs exist in many plant lineages (Figure 2B), suggesting the fundamental roles of MCTPs in plant development. The number of MCTPs is greatly expanded in dicotyledons and monocotyledons (Figure 2B), indicating that the family members of MCTPs among species increase substantially after several rounds of whole-genome duplication, and may evolve to generate functional specialized MCTPs to respond to changing environmental stimuli.
Expression Analysis of GhMCTPs in Upland Cotton
To identify the potential roles of GhMCTPs in cotton development, the expression patterns of all GhMCTPs were investigated in various cotton tissues, including root, stem, leaf, main stem apex, torus, calycle, pistil petal, and stamen, through analyzing previously published transcriptome datasets (You et al., 2017). We also analyzed the expression of all MCTPs in ovules and fibers at several developmental stages (Figure 3A). Based on their expression profiles, GhMCTPs were generally divided into two groups. One group of GhMCTPs was highly expressed in almost all tissues, including GhMCTP3-A/D, GhMCTP4-A/D, GhMCTP5-A/D, GhMCTP7-A/D, GhMCTP12-A/D, GhMCTP14-A/D, and GhMCTP17-A/D. Members of the other groups were tissue-preferred genes (Figure 3A). For example, GhMCTP1-A and GhMCTP2-A/D were preferentially expressed in petals. GhMCTP6-D, GhMCTP9-A/D, GhMCTP10-A/D, and GhMCTP16-A/D were highly expressed in the main stem apex and ovules at early stages (Figure 3A). This tissue/organ-preferred expression pattern indicates that these MCTPs may function in specific developmental stages.
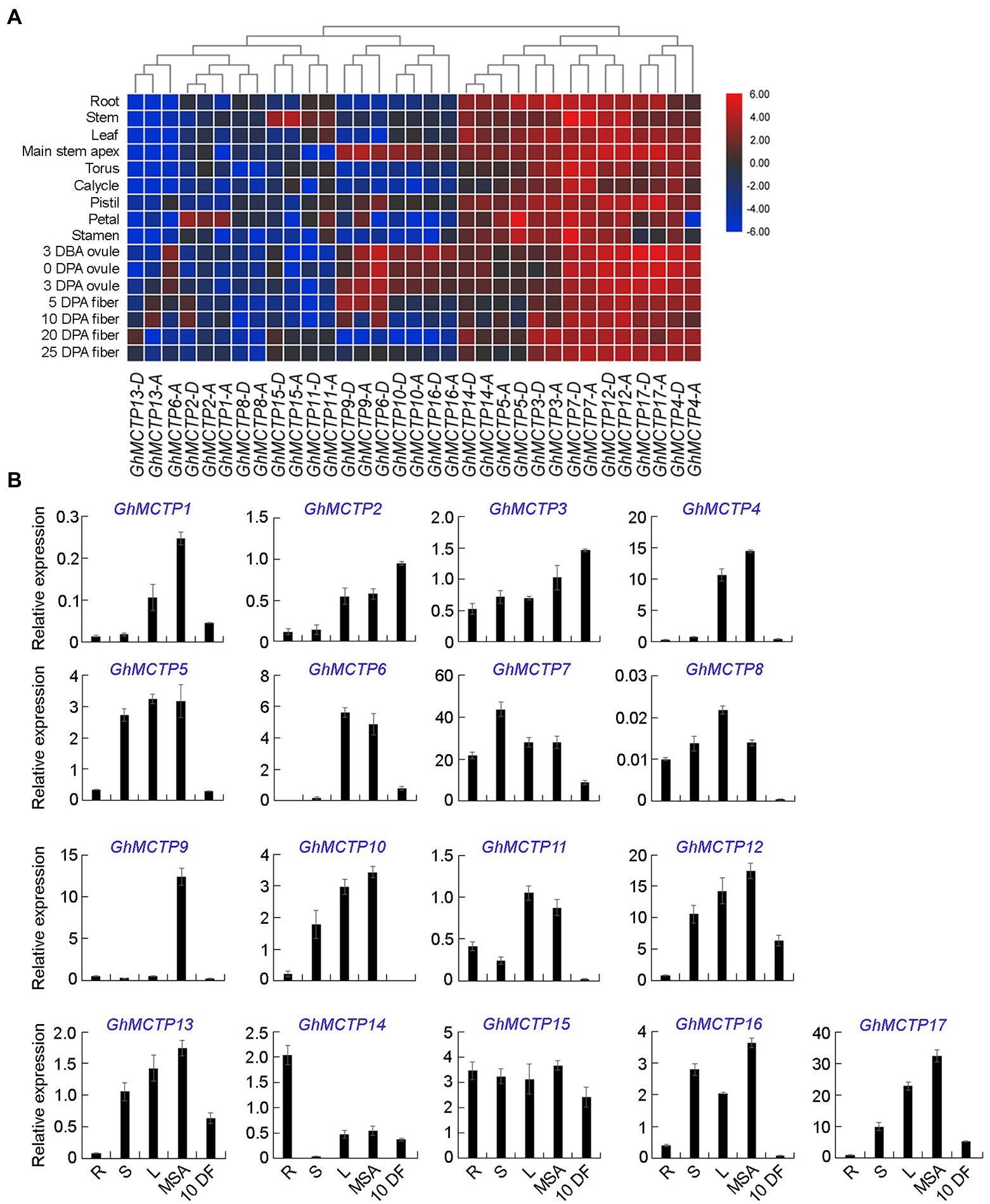
Figure 3. Expression patterns of GhMCTPs in upland cotton. (A) Heat map analysis of GhMCTP gene expressions in different organs of upland cotton. The relative fold changes in gene expression for all GhMCTP genes were compared. The color from blue to red indicates low to high expression. DBA, days before-anthesis; DPA, days post-anthesis. (B) Quantitative real-time PCR (qRT-PCR) analysis of 17 GhMCTPs in various tissues of upland cotton. Results were normalized against the expression level of GhUBI1. R, root; S, stem; L, leaf; MSA, main stem apex; and 10 DF, fibers at 10-day post-anthesis. Error bars indicate SD.
We further isolated different cotton tissues, including root, stem, leaf, main stem apex, and fibers at 10-day post-anthesis, and carried out qRT-PCR to investigate expression profiles of all GhMCTPs (Figure 3B). The results revealed that GhMCTP genes were expressed in all organs and tissues detected. Furthermore, we found that GhMCTPs from different subfamilies exhibited different expression patterns (Figure 3), suggesting that GhMCTPs might be involved in different cotton developmental processes.
GhMCTP7, GhMCTP12, and GhMCTP17 Function Additively to Regulate Cotton Shoot Meristem Development
The majority of GhMCTP genes were highly detected in the main stem apex (Figures 3A,B), suggesting their possible roles in meristem development. In Arabidopsis, two MCTP proteins, FT INTERACTING PROTEIN 3 (FTIP3) and FTIP4 have been reported to play an essential role in mediating shoot meristem development, thus determining the overall plant architecture (Liu et al., 2018b). GhMCTP12-A and GhMCTP12-D were the closest homologs of FTIP3 and FTIP4 in G. hirsutum (Figure 2A). In cotton, GhMCTP7-A/D and GhMCTP17-A/D showed sequence similarity with GhMCTP12-A/D (Figure 2A), and GhMCTP7, GhMCTP12, and GhMCTP17 were all highly expressed in the main stem apex (Figure 3). These results suggest that they are potential candidates to regulate meristem development.
The effect of different cotton cultivars on plant height was highly significant. We chose eight allotetraploid cotton cultivars and divided them into two groups based on their plant heights (Figure 4A). To understand the roles of GhMCTPs in shoot development, we examined the expressions of GhMCTP7, GhMCTP12, and GhMCTP17 in the shoot apex of selected allotetraploid cotton cultivars and found that the expression levels of GhMCTP7, GhMCTP12, and GhMCTP17 were correlated with their plant heights (Figures 4B–D).
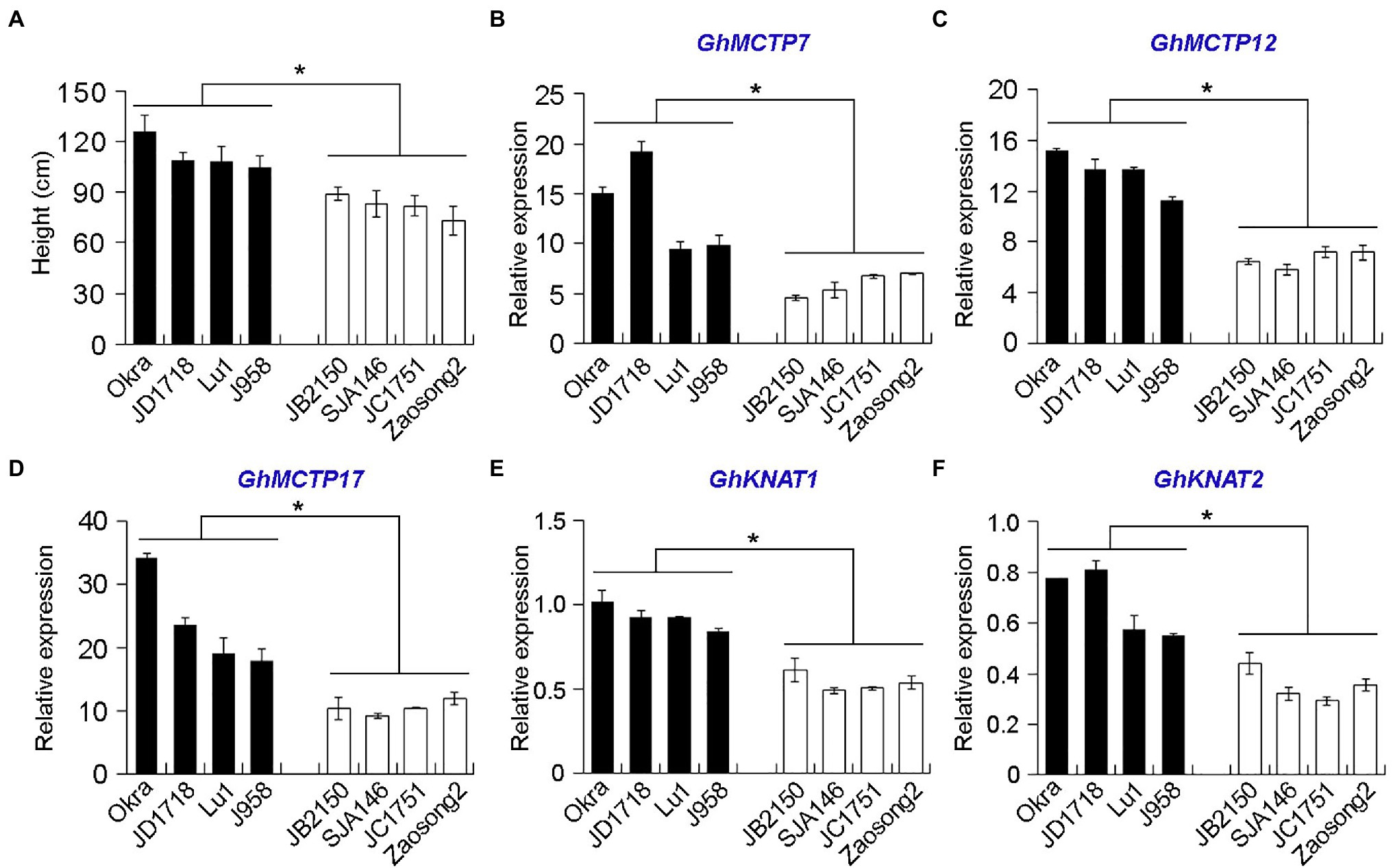
Figure 4. The expression levels of GhMCTP7/12/17 and GhKNAT1/2 are correlated with the average plant height (cm) of cotton varieties. (A) Comparison of plant height of eight allotetraploid cotton cultivars (G. hirsutun). The heights of plants are calculated when cotton bolls were opening. The cotton varieties are divided into two groups according to their plant heights. The varieties over 100cm in height or under 100cm are grouped and are shown in black bars or white bars, respectively. (B-F) Quantitative real-time PCR analysis of GhMCTP7 (B), GhMCTP12 (C), GhMCTP17 (D), GhKNAT1 (E), and GhKNAT2 (F) in main stem apex of different cotton species. The X-axis represents different upland cotton species, while the Y-axis represents gene relative expressions. Results were normalized against the expression level of GhUBI1. Error bars indicate SD of three biological replicates (*p<0.05).
Virus-induced gene silencing is a powerful reverse genetic technology for quick functional characterizations of plant genes, which serves as an alternative to mutant collection or creating stable transgenic lines (Gao et al., 2013; Lange et al., 2013; McGarry et al., 2016). To further understand the biological functions of GhMCTPs in shoot development, we silenced the gene expression of GhMCTP7, GhMCTP12, and GhMCTP17 using TRV-based VIGS technique. Real-time quantitative PCR assays showed that the expressions of GhMCTPs were downregulated in cotton main stem apex of TRV2:GhMCTP plants compared to negative control plants treated with TRV2:00 (Supplementary Figure 3); however, most of the VIGS-mediated gene silencing lines exhibited mild dwarf phenotype compared to those of negative control plants (Figures 5A–C). We reasoned that GhMCTP7, GhMCTP12, and GhMCTP17 might play a redundant role to regulate meristem development. Thus, we conducted a VIGS assay to simultaneously silence the expression of GhMCTP7, GhMCTP12, and GhMCTP17 (Supplementary Figure 3B). The resulting plants exhibited a dwarf phenotype, with down-curly leaves and occasionally shoot branching (Figure 5D; Supplementary Figure 4). Furthermore, the longitudinal sections showed that TRV2:GhMCTP7/12/17 plants had a narrow dome-shaped meristem, and the organs generated at the flanking of the meristems were also malformed and disordered positioned (Figures 5E–H). These results substantiate that GhMCTP7, GhMCTP12, and GhMCTP17 are essential for shoot meristem development.
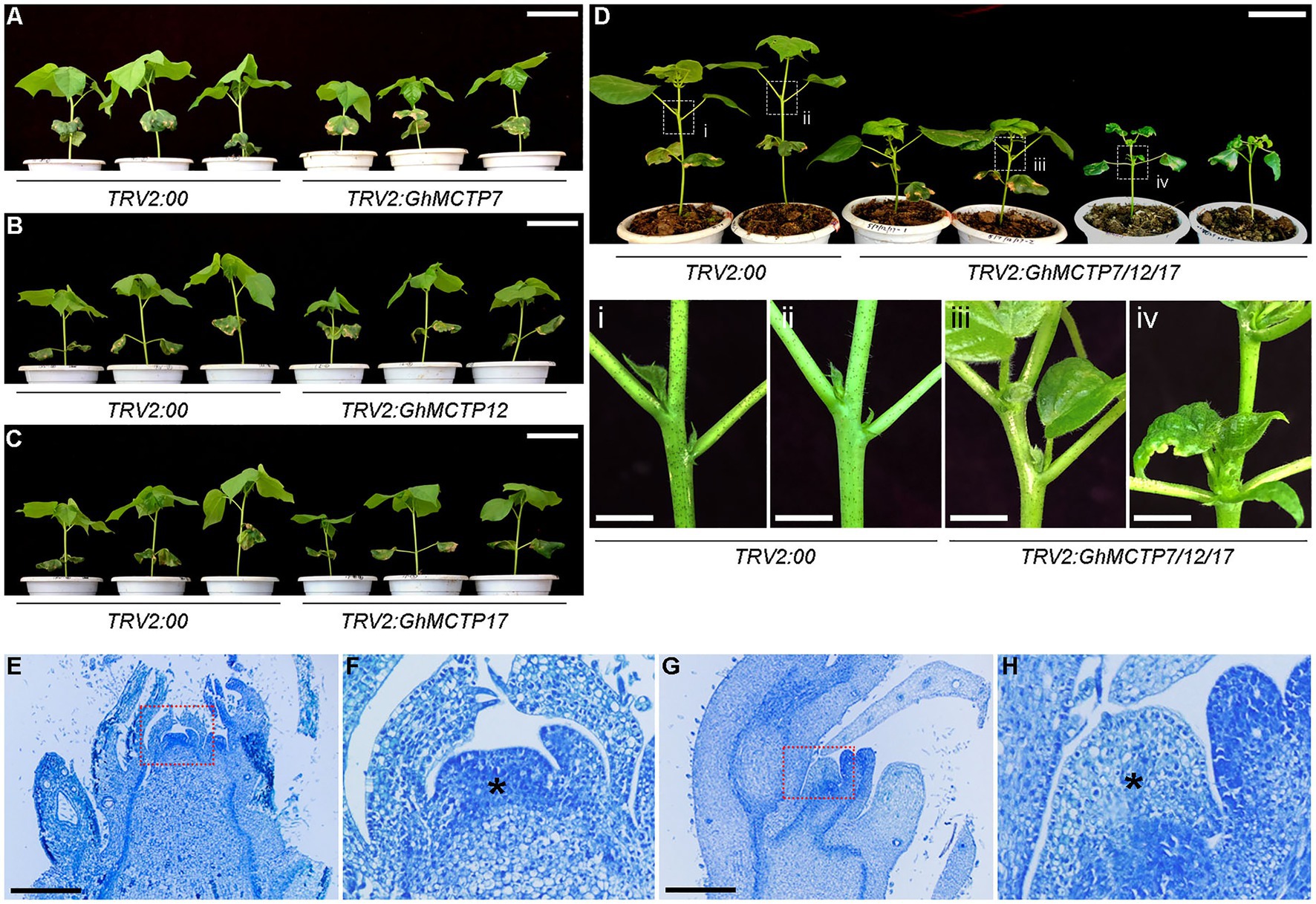
Figure 5. Phenotypic analyses of TRV2:GhMCTPs plants. (A–C) Plant height comparison of 3–4 leaf stage TRV2:GhMCTP7 (A), TRV2:GhMCTP12 (B), and TRV2:GhMCTP17 (C) plants with TRV2:00 plants (negative control group). Scale bars=5cm. (D) Plant height comparison of four leaf stage TRV2:GhMCTP7/12/17 with TRV2:00 plants (upper panel). The lower panels show the magnified view of boxes indicated in the upper panel. Scale bars=5cm (top) and 1cm (bottom). (E–H) Median longitudinal section of main inflorescence shoot apices of TRV2:00 (E) and TRV2:GhMCTP7/12/17 (G) plants. Scale bars=1μm. (F,H) The magnified views of boxes are indicated in (G) and (H), respectively. Asterisks indicate main inflorescence meristems.
Subcellular Localization of GhMCTP7, GhMCTP12, and GhMCTP17
To determine the subcellular localization of GhMCTP7, GhMCTP12, and GhMCTP17, we transiently expressed their full-length open reading frames fused with the green fluorescent protein (GFP)-MCTPs reporter in N. benthamiana leaf epidermal cells and observed two different types of subcellular localization patterns (Figure 6; Supplementary Figure 5). GhMCTP7 was localized to puncta-like structures in the cytosol within cells (Figure 6A; Supplementary Figure 5). We then coexpressed 35S:GFP-GhMCTP7 with the fluorescence-tagged endosome marker 35S:RFP-RabF2b (Jaillais et al., 2006), and observed GhMCTP7 was partially localized in endosomal compartments (Figure 6D). GhMCTP12 and GhMCTP17 were localized in whole cells and substantially colocalized with an ER marker, RFP-HDEL (Figures 6B,C,E; Nelson et al., 2007). These results suggest that GhMCTP7, GhMCTP12, and GhMCTP17 function coordinately to regulate intercellular signaling and control cotton development.
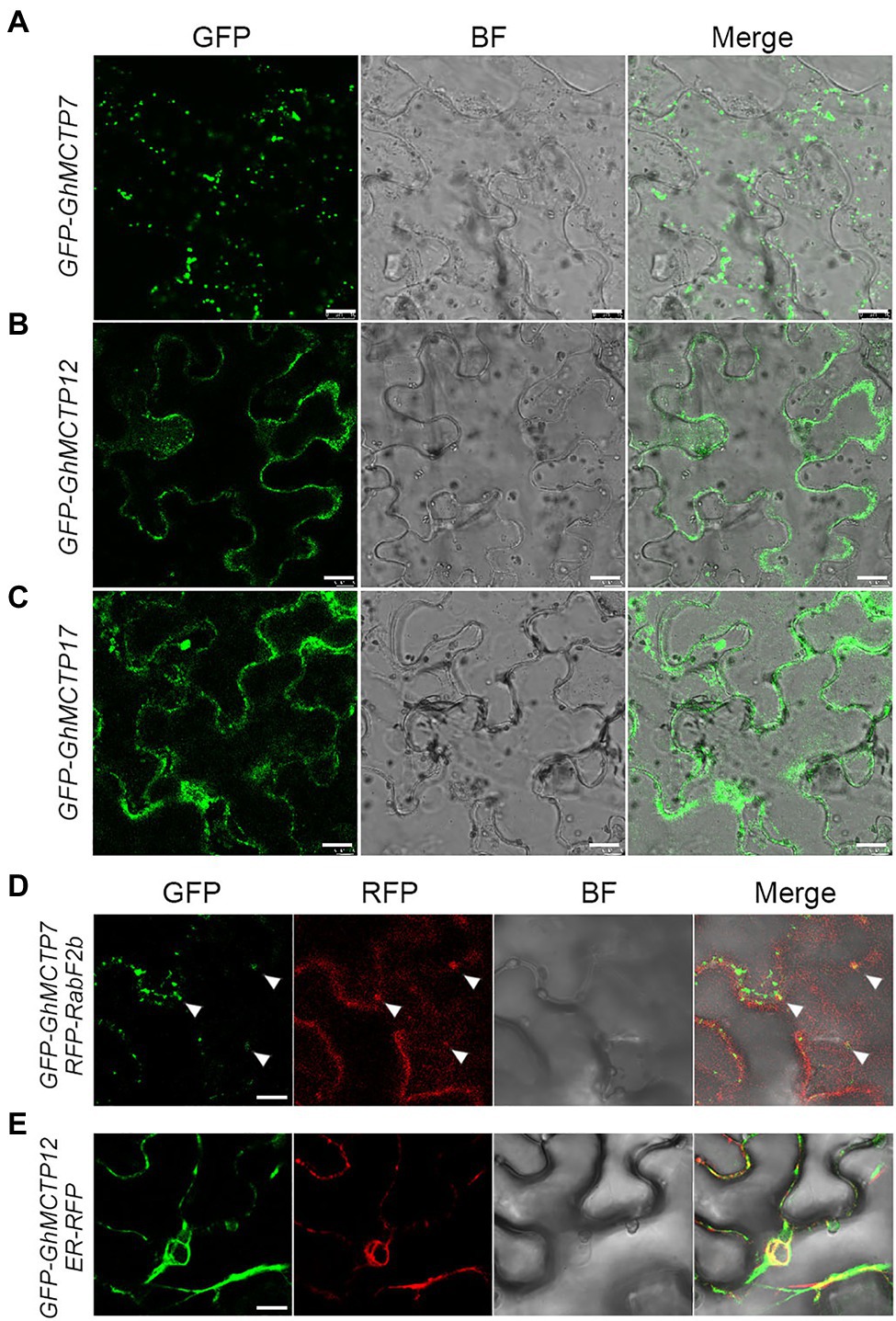
Figure 6. Subcellular localization of green fluorescent protein (GFP)-GhMCTP7/12/17 in Nicotiana benthamiana leaf epidermal cells. (A–C) Representative GFP fluorescence images of GFP-GhMCTP7 (A), GFP-GhMCTP12 (B), and GFP-GhMCTP17 (C). Scale bars=50μm. (D) Colocalization of GFP-GhMCTP7 with an endosome marker (RFP-RabF2b). (E) Co-expression of GFP-GhMCTP12 with an ER marker (ER-RFP). Scale bars=10μm.
GhMCTP7/12/17 Interact With GhKNAT1/2 to Regulate Shoot Development
FTIP3 and FTIP4, two MCTP proteins in Arabidopsis, are required for shoot apical meristem development through mediating subcellular localization and intercellular trafficking of STM (Liu et al., 2018b). GhMCTP7, GhMCTP12, and GhMCTP17 shared high sequence similarities with FTIP3 and FTIP4 in Arabidopsis (Figure 2A). Our finding on the tissue expression patterns of GhMCTP7, GhMCTP12, and GhMCTP17 in the shoot apex and their roles in cotton meristem development prompted us to investigate whether GhMCTP7, GhMCTP12, and GhMCTP17 interact with KNOTTED1 (KN1)-like homeobox (KNOX) family proteins in cotton, like their counterparts in Arabidopsis (Liu et al., 2018b).
First, we examined the expression profiles of all GhKNAT genes in different tissue of upland cotton. The results showed that GhKNAT1 and GhKNAT2 were highly expressed in the main stem apex (Supplementary Figure 6). We then investigated whether GhMCTP7/12/17 interact with GhKNAT1/2. We conducted a detailed analysis of protein interaction between GhMCTP7/12/17 and GhKNAT1/2. Yeast two-hybrid assays revealed that the GhMCTP7/12/17∆™, a truncated GhMCTP7/12/17 devoid of the transmembrane region, interacted with GhKNAT1/2 (Figure 7A). To test the interaction between GhMCTP7/12/17 and GhKNAT1/2 in planta, we performed luciferase complementation imaging (LCI) assays. We coexpressed nLUC-GhMCTP7/12/17 and cLUC-GhKNAT1/2 and detected fluorescence signals in N. benthamiana leaves (Figures 7B,C). These results demonstrate the protein interactions between GhMCTP7/12/17 and GhKNAT1/2 in plants.
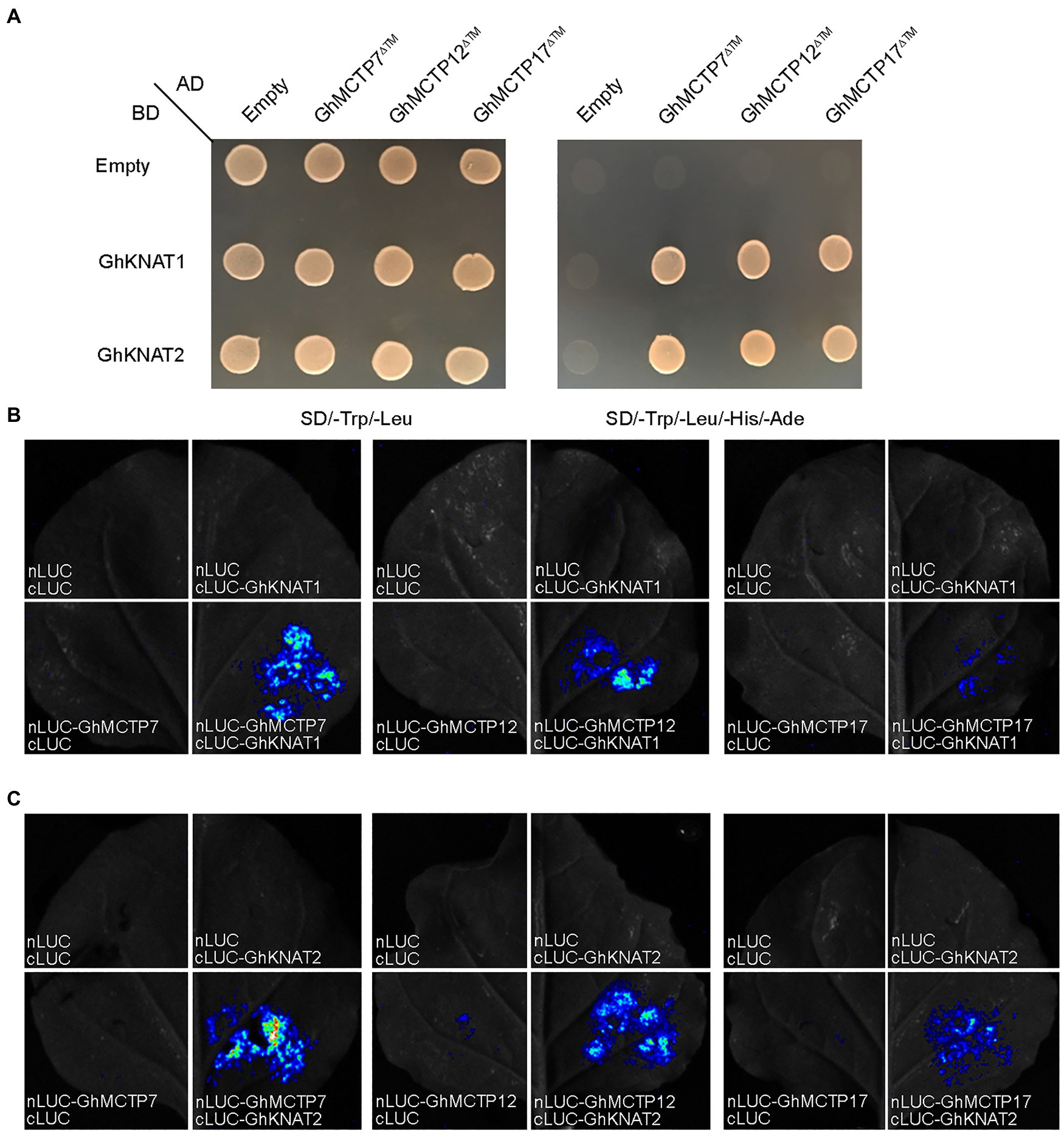
Figure 7. GhMCTP7/12/17 interact with GhKNAT1/2. (A) Yeast two-hybrid assay showing the interaction between GhMCTP7/12/17∆™ and GhKNAT1/2. Transformed yeast cells were grown on SD -Ade/-His/-Leu/-Trp medium. (B,C) Luciferase complementation imaging (LCI) assays showing that GhMCTP7/12/17 interact with GhKNAT1 (B) and GhKNAT2 (C) in N. benthamiana leaves. Fluorescence signal intensities represent their protein interaction intensities.
Comparing the expression levels of GhKNAT1 and GhKNAT2 in the shoot apex of selected allotetraploid cotton cultivars, we also found that the expression levels of GhKNAT1 and GhKNAT2 were higher in the taller cotton variants (Figures 4E,F), suggesting that GhKNAT1 and GhKNAT2 are the potential regulators for meristem development. Then, we further investigated the effects of GhKNAT1 and GhKNAT2 on meristem development. We silenced the expression of GhKNAT1 and GhKNAT2 using the VIGS in soil-grown upland cotton. Most of these TRV2:GhKNAT1/2 plants exhibited a dwarf phenotype (Figure 8A). Examination of some selected plants revealed that the expressions of GhKNAT1 and GhKNAT2 were downregulated in these TRV2:GhKNAT1/2 plants (Figure 8B), suggesting that GhKNAT1 and GhKNAT2 regulate meristem development.
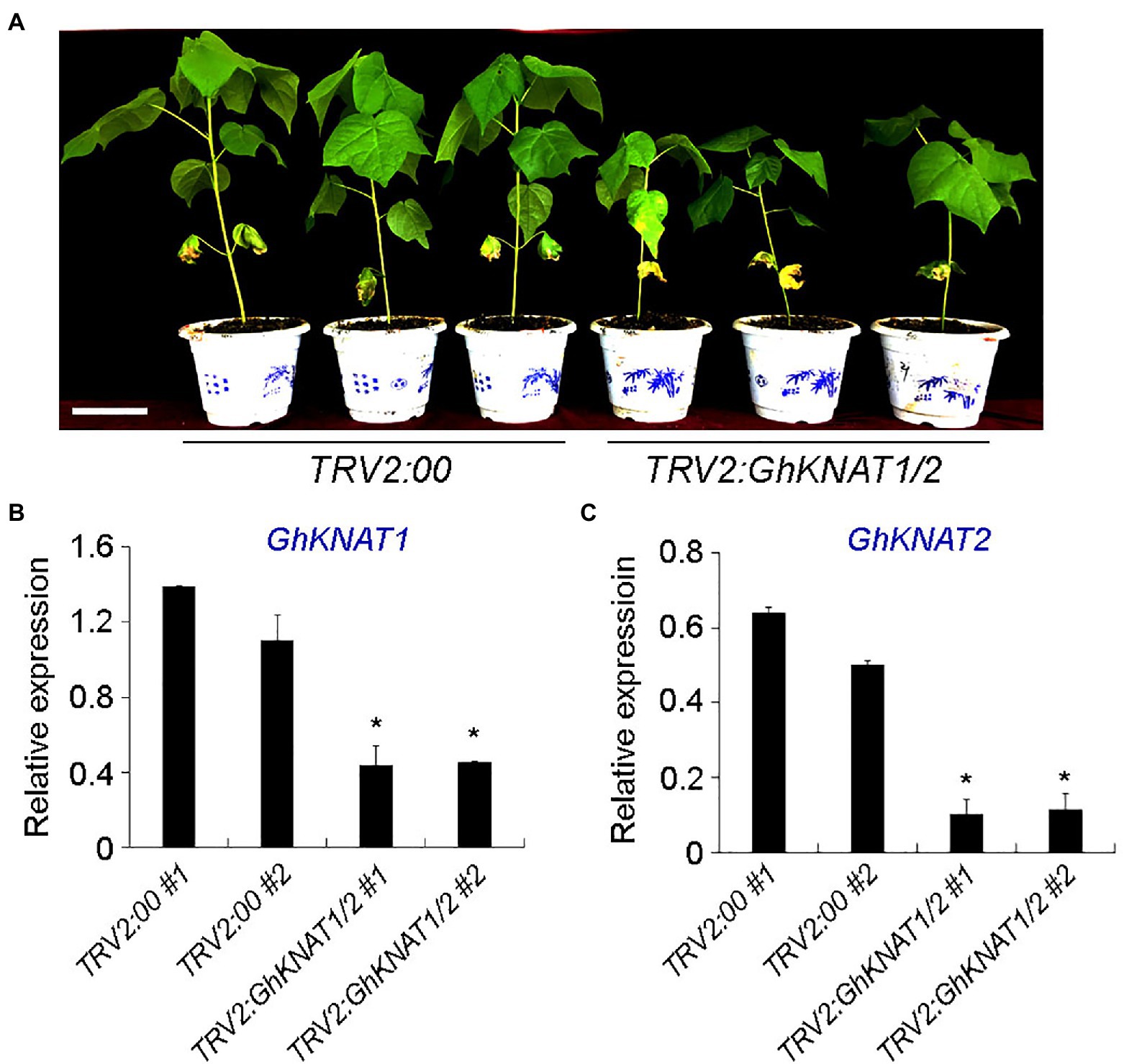
Figure 8. Phenotypic analyses of TRV2:GhKNAT1/2 plants. (A) Plant height comparison of 5–6 leaf stage TRV2:GhKNAT1/2 plants with TRV2:00 plants (negative control group). (B,C) Expression analysis of GhKNAT1 and GhKNAT2 in main stem apex of TRV2:00 (B) and TRV2:GhKNAT1/2 (C) plants, respectively. Results were normalized against the expression level of GhUBI1. Error bars indicate SD of three biological replicates (*p<0.05).
GhMCTP7/12/17 Are Involved in the Regulation of Multiple Signal Pathways
KNOTTED1-like homeobox transcription factors promote meristem function through directly targeting various transcription factors and genes participating in hormone pathways (Bolduc et al., 2012). Given that GhMCTP7/12/17 may regulate meristem development through mediating the function of GhKNAT1/2, we further conducted a detailed expression analysis to investigate the involvement of GhMCTP7/12/17 in regulating key regulators in meristem development and hormone signaling pathway.
To further characterize the functions of GhMCTP7/12/17 and GhKNAT1/2, the expressions of selected genes, which counterparts in Arabidopsis are key regulators in shoot meristem development and genes in various hormone signaling pathways, were analyzed. First, key regulators with high expression in the main stem apex were selected based on the public expression data (Supplementary Figures 7–10). We then carried out qRT-PCR to examine the expressions of the selected genes in the main stem apex of TRV2:00, TRV2:GhMCTP7/12/17, and TRV2:GhKNAT1/2 plants (Figure 9). Gh AGAMOUS-LIKE 8-1/-2 (GhAGL8-1/-2), the homologs of APETALA1 (AP1) in cotton, regulate plant height and early maturity of cotton (Su et al., 2018). GENERAL REGULATORY FACTORs (GRFs) play important roles in flowering regulation and meristem development (Sang et al., 2021). We found that the expressions of GhAGL8-1/2 and GhGRF6 were downregulated in both TRV2:GhMCTP7/12/17 and TRV2:GhKNAT1/2, suggesting the delayed determination of floral meristem identity (Figures 9A,B).
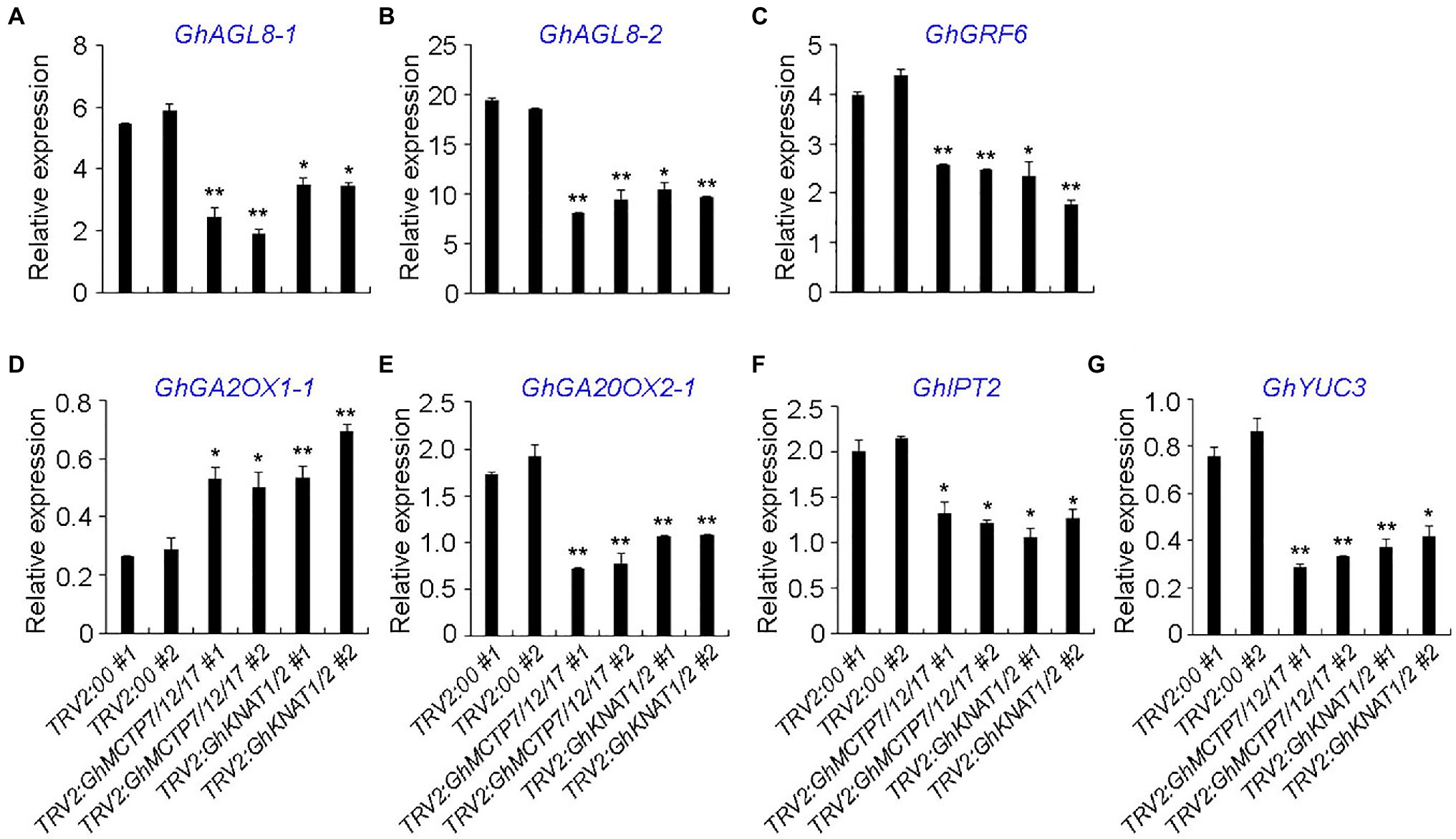
Figure 9. Quantitative analysis of multiple shoot meristem regulators in main stem apex of TRV2:00, TRV2:GhMCTP7/12/17, and TRV2:GhKNAT1/2 plants. The expression levels of GhALG8-1 (A), GhAGL8-2 (B), GhGRF6 (C), GhGA2OX1-1 (D), GhGA20OX2-1 (E), GhIPT2 (F), and GhYUC3 (G) in TRV2:GhMCTP7/12/17 and TRV2:GhKNAT1/2 plants. Results were normalized against the expression level of GhUBI1. Error bars indicate SD of three biological replicates (*p<0.05; **p<0.01).
KNOTTED1-like homeobox protein promotes meristem development through repressing gibberellin (GA) biosynthesis and activating cytokinin (CK) pathway (Jasinski et al., 2005). KNOX protein also regulates the expressions of auxin-related genes (Bolduc et al., 2012), suggesting that KNOX protein integrates multiple phytohormone signaling pathways to regulate meristem development. We studied the expression of cytokinin biosynthesis and signaling genes, auxin biosynthesis and signaling genes, GA biosynthesis and catabolism genes in both TRV2:GhMCTP7/12/17 and TRV2:GhKNAT1/2. We observed that the expression of GA catabolic enzyme GhGA2OX1-1 was elevated in both TRV2:GhMCTP7/12/17 and TRV2:GhKNAT1/2, whereas GA biosynthesis gene GhGA20OX2-1, CK biosynthesis gene Gh ISOPENTENYLTRANSFERASE 1 (GhIPT1), and auxin biosynthesis gene Gh YUCCA3 (GhYUC3) were consistently downregulated in TRV2:GhMCTP7/12/17 and TRV2:GhKNAT1/2 (Figures 9E–G). These results suggest that MCTP7/12/17 regulate meristem development partially through GhKNAT1/2-mediated regulatory pathway.
We also observed that some other regulators in meristem development and phytohormone signaling pathway in main stem apex of TRV2:00, TRV2:GhMCTP7/12/17, and TRV2:GhKNAT1/2 plants (Supplementary Figure 11). Stem cell regulators Gh AINTEGUMENTA-1/-2/-3 (GhANT-1/-2/-3), Gh GROWTH-REGULATING FACTOR 1 (GhGRF1), and Gh GRF1-INTERACTING FACTOR 3 (GhGIF3) were downregulated in TRV2:GhMCTP7/12/17 plants (Mudunkothge and Krizek, 2012; Kim and Tsukaya, 2015); however, their expressions in TRV2:GhKNAT1/2 plants were not changed (Supplementary Figures 11A–E). In addition, we observed that, although the expressions of Gh CLAVATA3/ESR-RELATED 27 (GhCLE27) and Gh KNOTTED1-LIKE HOMEOBOX GENE 4 (GhKNAT4) were consistently downregulated in both TRV2:GhMCTP7/12/17 and TRV2:GhKNAT1/2 plants, the downregulation of GhKNAT4 and GhCLE27 was more obvious in TRV2:GhMCTP7/12/17 plants (Fletcher, 2020; Supplementary Figures 11F,G). Auxin efflux regulator Gh PIN-FORMED 3/-2 (GhPIN3/-2) and Gh PHOTOSYSTEM I LIGHT HARVESTING COMPLEX GENE2 (Gh LHCA2) has been reported to regulate cotton height (Su et al., 2018; Ma et al., 2019). We observed that the expressions of GhPIN3/-2 and GhLHCA2 were downregulated in TRV2:GhMCTP7/12/17 plants, but minor altered in TRV2:GhKNAT1/2 plants (Supplementary Figures 11H–J). Furthermore, the expression of GhARR5D, which functions in the cytokinin signaling pathway, was increased in TRV2:GhMCTP7/12/17 plants but not in TRV2:GhKNAT1/2 plants (Supplementary Figure 11L). These results demonstrate that GhMCTP7, GhMCTP12, and GhMCTP17 also regulate meristem development independent of GhKNAT1 and GhKNAT2, possible through other KNOX family members.
Discussion
The development of multicellular organisms relies on the coordination of a variety of specialized cell types through intercellular communication. MCTP family proteins in Arabidopsis and its orthologs in several plant species have been shown to play an important role in protein intercellular movement and are essential for plant development (Liu et al., 2013, 2019; Hao et al., 2020; Zhu et al., 2020). MCTP proteins have been identified in the cotton genome (Hao et al., 2020); however, their biological functions are still largely unknown. In this study, we identified 33 GhMCTP genes from the upland cotton genome and analyzed their evolutionary relationships. Through examining the expression patterns of all GhMCTPs in different tissues of upland cotton, we found that GhMCTP7, GhMCTP12, and GhMCTP17 are highly expressed in the main stem apex and play a key role in shoot development. GhMCTP7/12/17 interacted with GhKNAT1/2 and modulated the expression of multiple shoot meristem regulators in a GhKNAT1/2-dependent and independent manner. Our findings suggest that GhMCTP proteins are evolutionarily conserved in upland cotton and play conserved roles in meristem development.
We have systematically characterized 33 GhMCTP genes in G. hirsutum, which are grouped into seven clades based on the phylogenetic analysis (Figure 1A). The key feature of MCTPs is the presence of multiple C2 domain at the N-terminus and PRT_C domain at the C-terminus (Figure 1C). All MCTPs in Arabidopsis contain the C-terminal transmembrane region, which anchors the MCTP in the intracellular membrane. In upland cotton, GhMCTP3 does not contain the C-terminal transmembrane region, and the transmembrane regions of some GhMCTPs are located in the N-terminal C2 domain region (Supplementary Figure 2), suggesting the functional divergence among MCTPs in different species. Comparing with the limited number of MCTPs in animals, a large number of MCTPs are identified in cotton and other plant lineages (Figure 2A), implying that plants have evolved functional specialized MCTPs to regulate distinct biological processes. We also noticed that all GhMCTPs exist in cotton A subgenome and D subgenome, demonstrating their fundamental roles in cotton development. Chromosome distribution of GhMCTP shows that GhMCTP genes are dispersed across the chromosome but not in a cluster pattern (Supplementary Figure 1), indicating that the GhMCTP gene family does not simply arise from chromosome region duplication but also are involved in the extensive reshuffling and divergent evolution. It is also noteworthy that MCTP family proteins exist in most plant lineages (Figure 2B), suggesting the fundamental roles of MCTPs in plant development. GhMCTPs exhibit distinct or overlapping expression patterns in various tissues at different developmental stages (Figure 3), demonstrating MCTPs are functionally specialized during plant evolution.
In Arabidopsis, different MCTPs showed distinct patterns in various tissues, and no MCTPs exhibited the identical expression pattern at both vegetative and reproductive tissues (Liu et al., 2018a), suggesting that MCTPs might be differentially regulated and play different roles in various tissues. Additionally, some MCTPs share similar expression patterns in several tissues (Liu et al., 2018a), indicating that MCTPs might function redundantly during plant development. To understand the biological function of GhMCTPs in cotton, we examined the expression patterns of all GhMCTP in various tissues and revealed their distinct or overlapping expressions in various tissues (Figure 3). GhMCTP7, GhMCTP12, and GhMCTP17 are highly expressed in the shoot apex, and their expression levels are correlated with the plant heights in different allotetraploid cotton cultivars. Furthermore, plants with the downregulation of GhMCTP7, GhMCTP12, and GhMCTP17 exhibit dwarf phenotype, demonstrating that they might function redundantly to regulate meristem development.
Multiple C2 domain and transmembrane region proteins have been shown to regulate multiple developmental processes by mediating the trafficking of various macromolecules. In Arabidopsis, FTIP3 and FTIP4 interact with and regulate STM intercellular and intracellular trafficking, thus affecting the protein distribution within the meristem (Liu et al., 2018b). Regulation of STM trafficking at subcellular and tissue levels causes early termination of shoot apices and continuously generation secondary shoots, resulting in dwarf and bushy phenotypes. The position of leaves and branches, timing of the flowering, and relative position of reproductive structures are traits that affect cotton productivity. Genetic engineering the cotton architecture is crucial for cotton domestication and will benefit crop production. GhMCTP7, GhMCTP12, and GhMCTP17 show high sequence similarity with FTIP3 and FTIP4, hinting that GhMCTP7, GhMCTP12, and GhMCTP17 might regulate meristem development through KNOX family proteins. Considering the protein interactions between GhMCTP7/12/17 and GhKNAT1/2 (Figures 7, 8), we reasoned that GhMCTP7, GhMCTP12, and GhMCTP17 might regulate the function of GhKNAT1 and GhKNAT2 to modulate meristem development. Consistently, gene silencing of GhKNAT1 and GhKNAT2 in TRV2:GhKNAT1/2 results in a dwarf phenotype (Figure 7), similar to TRV2:GhMCTP7/12/17.
KNOTTED1-like homeobox-like transcription factors promote meristem function through regulating various transcription factors and manipulating phytohormone pathways (Jasinski et al., 2005; Bolduc et al., 2012). We examined some of the putative downstream targets of GhKNAT1 and GhKNAT2 in the main apex of TRV2:00, TRV2:GhMCTP7/12/17, and TRV2:GhKNAT1/2 plants (Figure 9). The expression levels of several GhKNAT1/2 putative targets are consistently upregulated or downregulated in both TRV2:GhMCTP7/12/17 and TRV2:GhKNAT1/2 plants (Figure 9). We also observed that genes with altered expression in TRV2:GhMCTP7/12/17 are not similarly affected in TRV2:GhKNAT1/2, suggesting that GhMCTP7, GhMCTP12, and GhMCTP17 also regulate meristem development independent of GhKNAT1 and GhKNAT2 (Supplementary Figure 11). Taken together, these results demonstrate that GhMCTP7, GhMCTP12, and GhMCTP17 function redundantly to regulate meristem development partially through GhKNAT1 and GhKNAT2.
Plant development requires cell-cell communication and the coordination of various specialized cell types. Non-cell-autonomous signals are one of the regulatory mechanisms to integrate various signals and coordinate plant development. Through the combination of different approaches, an increasing number of mobile signals, such as transcription factors and peptides, have been discovered to regulate plant growth. Severe mutations of key regulators in plants always lead to embryonic lethality or obvious growth defect, which precludes evaluation of later phenotypes and prevents its application for crop breeding (Paaby and Rockman, 2013). Genetic engineering the cis-elements of key regulators will bypass its lethality effect and expose multiple pleiotropic roles of this gene (Hendelman et al., 2021). Regulation of protein trafficking will be another alternative approach to make dysfunction of key regulators. MCTP-mediated regulation of proteins at sub-cellular and tissue levels provides a chance to modulate the protein function at the desired degree, which could generate a phenotype that different from the severe mutants and provide important implications for biotechnological application for crop breeding.
Conclusion
In conclusion, our systematic analysis of GhMCTPs in upland cotton illustrates their diverse expression patterns. We find that GhMCTP7, GhMCTP12, and GhMCTP17 are highly expressed in the main stem apex, and they function redundantly to regulate the development of the main stem apex and affect cotton architecture. We also show that the expression levels of GhMCTP7, GhMCTP12, and GhMCTP17 in the shoot apex of eight selected allotetraploid cotton cultivars are correlated with their plant heights. In addition, VIGS plants silenced for GhMCTP7, GhMCTP12, and GhMCTP17 show a dwarf phenotype. Taken together, this study provides important clues for studying the function of GhMCTPs, deepens our understanding of cotton apex regulation, and establishes a resource for cotton breeding.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
GH, QH, and MZ conceived and designed the experiment and performed most of the experiments. MW, XH, and JL performed some of the experiments and assisted in data analysis. CF analyzed some data. QH, LX, LL, and GH analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 31271317), Outstanding Youth Science Fund of Xinjiang Uygur Autonomous Region (grant no. 2021D01E17), Fundamental Research Funds for the Central Universities (grant nos. CCNU16A02047 and CCNU19TS064), and the Shanghai Pujiang Program (20PJ1405200).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the Germplasm Bank of Cotton Institute of Shanxi Academy of Agricultural Sciences for providing the cotton seeds (Zaosong 2).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.767667/full#supplementary-material
Footnotes
1. ^https://www.cottongen.org/blast/protein/protein
2. ^http://myhits.isb-sib.ch/cgi-bin/motif_scan
3. ^http://www.ebi.ac.uk/Tools/pfa/iprscan
4. ^http://smart.emblheidelberg.de/
5. ^https://www.cottongen.org/find/genes
6. ^http://us.expasy.org/tools/protparam.html
7. ^https://www.cottongen.org/
8. ^https://phytozome.jgi.doe.gov/pz/portal.html
9. ^https://www.megasoftware.net
10. ^http://gsds.cbi.pku.edu.cn/
11. ^https://www.cottongen.org/species/Gossypium_hirsutum/ZJU-AD1_v2.1
References
Bolduc, N., Yilmaz, A., Mejia-Guerra, M. K., Morohashi, K., O'Connor, D., Grotewold, E., et al. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26, 1685–1690. doi: 10.1101/gad.193433.112
Brault, M. L., Petit, J. D., Immel, F., Nicolas, W. J., Glavier, M., Brocard, L., et al. (2019). Multiple C2 domains and transmembrane region proteins (MCTPs) tether membranes at plasmodesmata. EMBO Rep. 20:e47182. doi: 10.15252/embr.201847182
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y., Shen, J., Zhang, L., Qi, H., Yang, L., Wang, H., et al. (2021). Nuclear translocation of OsMFT1 that is impeded by OsFTIP1 promotes drought tolerance in rice. Mol. Plant 14, 1297–1311. doi: 10.1016/j.molp.2021.05.001
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 323–324. doi: 10.1104/pp.107.111740
Cho, W., and Stahelin, R. V. (2006). Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta 1761, 838–849. doi: 10.1016/j.bbalip.2006.06.014
Corbalan-Garcia, S., and Gómez-Fernández, J. C. (2014). Signaling through C2 domains: more than one lipid target. Biochim. Biophys. Acta 1838, 1536–1547. doi: 10.1016/j.bbamem.2014.01.008
Fletcher, J. C. (2020). Recent advances in Arabidopsis CLE peptide signaling. Trends Plant Sci. 25, 1005–1016. doi: 10.1016/j.tplants.2020.04.014
Gao, W., Long, L., Zhu, L.-F., Xu, L., Gao, W.-H., Sun, L.-Q., et al. (2013). Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteomics 12, 3690–3703. doi: 10.1074/mcp.M113.031013
Genç, Ö., Dickman, D. K., Ma, W., Tong, A., Fetter, R. D., and Davis, G. W. (2017). MCTP is an ER-resident calcium sensor that stabilizes synaptic transmission and homeostatic plasticity. eLife 6:e22904. doi: 10.7554/eLife.22904
Hao, P., Wang, H., Ma, L., Wu, A., Chen, P., Cheng, S., et al. (2020). Genome-wide identification and characterization of multiple C2 domains and transmembrane region proteins in Gossypium hirsutum. BMC Genomics 21:445. doi: 10.1186/s12864-020-06842-1
Hendelman, A., Zebell, S., Rodriguez-Leal, D., Dukler, N., Robitaille, G., Wu, X., et al. (2021). Conserved pleiotropy of an ancient plant homeobox gene uncovered by cis-regulatory dissection. Cell 184, 1724.e16–1739.e16. doi: 10.1016/j.cell.2021.02.001
Huang, G.-Q., Gong, S.-Y., Xu, W.-L., Li, W., Li, P., Zhang, C.-J., et al. (2013). A fasciclin-like arabinogalactan protein, GhFLA1, is involved in fiber initiation and elongation of cotton. Plant Physiol. 161, 1278–1290. doi: 10.1104/pp.112.203760
Huang, G., Wu, Z., Percy, R. G., Bai, M., Li, Y., Frelichowski, J. E., et al. (2020). Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat. Genet. 52, 516–524. doi: 10.1038/s41588-020-0607-4
Jaillais, Y., Fobis-Loisy, I., Miège, C., Rollin, C., and Gaude, T. (2006). AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443, 106–109. doi: 10.1038/nature05046
Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, I., et al. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15, 1560–1565. doi: 10.1016/j.cub.2005.07.023
John, M. E., and Crow, L. J. (1992). Gene expression in cotton (Gossypium hirsutum L.) fiber: cloning of the mRNAs. Proc. Natl. Acad. Sci. U. S. A. 89, 5769–5773. doi: 10.1073/pnas.89.13.5769
Kim, J. H., and Tsukaya, H. (2015). Regulation of plant growth and development by the growth-regulating factor and GRF-interacting factor duo. J. Exp. Bot. 66, 6093–6107. doi: 10.1093/jxb/erv349
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lange, M., Yellina, A. L., Orashakova, S., and Becker, A. (2013). Virus-induced gene silencing (VIGS) in plants: an overview of target species and the virus-derived vector systems. Methods Mol. Biol. 975, 1–14. doi: 10.1007/978-1-62703-278-0_1
Lek, A., Evesson, F. J., Sutton, R. B., North, K. N., and Cooper, S. T. (2012). Ferlins: regulators of vesicle fusion for auditory neurotransmission, receptor trafficking and membrane repair. Traffic 13, 185–194. doi: 10.1111/j.1600-0854.2011.01267.x
Liu, L., Li, C., Liang, Z., and Yu, H. (2018a). Characterization of multiple C2 domain and transmembrane region proteins in Arabidopsis. Plant Physiol. 176, 2119–2132. doi: 10.1104/pp.17.01144
Liu, L., Li, C., Song, S., Teo, Z. W. N., Shen, L., Wang, Y., et al. (2018b). FTIP-dependent STM trafficking regulates shoot meristem development in Arabidopsis. Cell Rep. 23, 1879–1890. doi: 10.1016/j.celrep.2018.04.033
Liu, L., Li, C., Teo, Z. W. N., Zhang, B., and Yu, H. (2019). The MCTP-SNARE complex regulates florigen transport in Arabidopsis. Plant Cell 31, 2475–2490. doi: 10.1105/tpc.18.00960
Liu, L., Liu, C., Hou, X., Xi, W., Shen, L., Tao, Z., et al. (2012). FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 10:e1001313. doi: 10.1371/journal.pbio.1001313
Liu, L., Zhu, Y., Shen, L., and Yu, H. (2013). Emerging insights into florigen transport. Curr. Opin. Plant Biol. 16, 607–613. doi: 10.1016/j.pbi.2013.06.001
Ma, J., Pei, W., Ma, Q., Geng, Y., Liu, G., Liu, J., et al. (2019). QTL analysis and candidate gene identification for plant height in cotton based on an interspecific backcross inbred line population of Gossypium hirsutum×Gossypium barbadense. Theor. Appl. Genet. 132, 2663–2676. doi: 10.1007/s00122-019-03380-7
Maeda, I., Kohara, Y., Yamamoto, M., and Sugimoto, A. (2001). Large-scale analysis of gene function in caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11, 171–176. doi: 10.1016/S0960-9822(01)00052-5
McGarry, R. C., Prewitt, S. F., Culpepper, S., Eshed, Y., Lifschitz, E., and Ayre, B. G. (2016). Monopodial and sympodial branching architecture in cotton is differentially regulated by the Gossypium hirsutum SINGLE FLOWER TRUSS and SELF-PRUNING orthologs. New Phytol. 212, 244–258. doi: 10.1111/nph.14037
Mergner, J., Frejno, M., List, M., Papacek, M., Chen, X., Chaudhary, A., et al. (2020). Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 579, 409–414. doi: 10.1038/s41586-020-2094-2
Mudunkothge, J. S., and Krizek, B. A. (2012). Three Arabidopsis AIL/PLT genes act in combination to regulate shoot apical meristem function. Plant J. 71, 108–121. doi: 10.1111/j.1365-313X.2012.04975.x
Nalefski, E. A., and Falke, J. J. (1996). The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 5, 2375–2390. doi: 10.1002/pro.5560051201
Nelson, B. K., Cai, X., and Nebenführ, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x
Paaby, A. B., and Rockman, M. V. (2013). The many faces of pleiotropy. Trends Genet. 29, 66–73. doi: 10.1016/j.tig.2012.10.010
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Reinhardt, D., and Kuhlemeier, C. (2002). Plant architecture. EMBO Rep. 3, 846–851. doi: 10.1093/embo-reports/kvf177
Sang, N., Liu, H., Ma, B., Huang, X., Zhuo, L., and Sun, Y. (2021). Roles of the 14-3-3 gene family in cotton flowering. BMC Plant Biol. 21:162. doi: 10.1186/s12870-021-02923-9
Shilo, B.-Z., and Schejter, E. D. (2011). Regulation of developmental intercellular signalling by intracellular trafficking. EMBO J. 30, 3516–3526. doi: 10.1038/emboj.2011.269
Shin, O. H., Han, W., Wang, Y., and Sudhof, T. C. (2005). Evolutionarily conserved multiple C2 domain proteins with two transmembrane regions (MCTPs) and unusual Ca2+ binding properties. J. Biol. Chem. 280, 1641–1651. doi: 10.1074/jbc.M407305200
Song, S., Chen, Y., Liu, L., See, Y. H. B., Mao, C., Gan, Y., et al. (2018). OsFTIP7 determines auxin-mediated anther dehiscence in rice. Nat. Plants 4, 495–504. doi: 10.1038/s41477-018-0175-0
Song, S., Chen, Y., Liu, L., Wang, Y., Bao, S., Zhou, X., et al. (2017). OsFTIP1-mediated regulation of florigen transport in rice is negatively regulated by the ubiquitin-like domain kinase OsUbDKgamma4. Plant Cell 29, 491–507. doi: 10.1105/tpc.16.00728
Song, J. H., Kwak, S.-H., Nam, K. H., Schiefelbein, J., and Lee, M. M. (2019). QUIRKY regulates root epidermal cell patterning through stabilizing SCRAMBLED to control CAPRICE movement in Arabidopsis. Nat. Commun. 10:1744. doi: 10.1038/s41467-019-09715-8
Su, J., Li, L., Zhang, C., Wang, C., Gu, L., Wang, H., et al. (2018). Genome-wide association study identified genetic variations and candidate genes for plant architecture component traits in Chinese upland cotton. Theor. Appl. Genet. 131, 1299–1314. doi: 10.1007/s00122-018-3079-5
Tran, T. M., McCubbin, T. J., Bihmidine, S., Julius, B. T., Baker, R. F., Schauflinger, M., et al. (2019). Maize carbohydrate partitioning defective33 encodes an MCTP protein and functions in sucrose export from leaves. Mol. Plant 12, 1278–1293. doi: 10.1016/j.molp.2019.05.001
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Trehin, C., Schrempp, S., Chauvet, A., Berne-Dedieu, A., Thierry, A. M., Faure, J. E., et al. (2013). QUIRKY interacts with STRUBBELIG and PAL OF QUIRKY to regulate cell growth anisotropy during Arabidopsis gynoecium development. Development 140, 4807–4817. doi: 10.1242/dev.091868
Tunstall, N. E., Herr, A., de Bruyne, M., and Warr, C. G. (2012). A screen for genes expressed in the olfactory organs of Drosophila melanogaster identifies genes involved in olfactory behaviour. PLoS One 7:e35641. doi: 10.1371/journal.pone.0035641
Vaddepalli, P., Herrmann, A., Fulton, L., Oelschner, M., Hillmer, S., Stratil, T. F., et al. (2014). The C2-domain protein QUIRKY and the receptor-like kinase STRUBBELIG localize to plasmodesmata and mediate tissue morphogenesis in Arabidopsis thaliana. Development 141, 4139–4148. doi: 10.1242/dev.113878
Van Norman, J. M., Breakfield, N. W., and Benfey, P. N. (2011). Intercellular communication during plant development. Plant Cell 23, 855–864. doi: 10.1105/tpc.111.082982
Wang, Y., Liu, L., Song, S., Li, Y., Shen, L., and Yu, H. (2017). DOFT and DOFTIP1 affect reproductive development in the orchid Dendrobium Chao Praya smile. J. Exp. Bot. 68, 5759–5772. doi: 10.1093/jxb/erx400
You, Q., Xu, W., Zhang, K., Zhang, L., Yi, X., Yao, D., et al. (2017). ccNET: database of co-expression networks with functional modules for diploid and polyploid Gossypium. Nucleic Acids Res. 45, D1090–D1099. doi: 10.1093/nar/gkw910
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Keywords: cotton, main stem apex development, GhMCTP, gene expression, protein interaction, KNOX family protein
Citation: Hu Q, Zeng M, Wang M, Huang X, Li J, Feng C, Xuan L, Liu L and Huang G (2021) Family-Wide Evaluation of Multiple C2 Domain and Transmembrane Region Protein in Gossypium hirsutum. Front. Plant Sci. 12:767667. doi: 10.3389/fpls.2021.767667
Edited by:
Hai Du, Southwest University, ChinaReviewed by:
Yang Zhu, Zhejiang University, ChinaHantao Wang, Cotton Research Institute, Chinese Academy of Agricultural Sciences (CAAS), China
Zuo Ren Yang, Institute of Cotton Research, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2021 Hu, Zeng, Wang, Huang, Li, Feng, Xuan, Liu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gengqing Huang, gqhuang@mail.ccnu.edu.cn; Lu Liu, lu.liu@sjtu.edu.cn
†These authors have contributed equally to this work
 Qianqian Hu1†
Qianqian Hu1† Lu Liu
Lu Liu Gengqing Huang
Gengqing Huang