- 1Student Research Committee, School of Medicine, Iran University of Medical Sciences, Tehran, Iran
- 2Biotechnology Department, Faculty of Chemistry, University of Kashan, Kashan, Iran
- 3Proteomics Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 4Department of Gynecology and Obstetrics, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 5School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
- 6School of Medicine, Kashan University of Medical Sciences, Kashan, Iran
- 7Student Research Committee, Kashan University of Medical Sciences, Kashan, Iran
- 8Laser Research Centre, Faculty of Health Science, University of Johannesburg, Doornfontein, South Africa
- 9Biotechnology Research Center, Pharmaceutical Technology Institute, Mashhad University of Medical Sciences, Mashhad, Iran
- 10School of Pharmacy, Mashhad University of Medical Sciences, Mashhad, Iran
- 11Shahid Beheshti Fertility Clinic, Department of Gynecology and Obsteterics, Isfahan University of Medical Sciences, Isfahan, Iran
Gynecologic cancer is a significant cause of death in women worldwide, with cervical cancer, ovarian cancer, and endometrial cancer being among the most well-known types. The initiation and progression of gynecologic cancers involve a variety of biological functions, including angiogenesis and metastasis—given that death mostly occurs from metastatic tumors that have invaded the surrounding tissues. Therefore, understanding the molecular pathways underlying gynecologic cancer metastasis is critical for enhancing patient survival and outcomes. Recent research has revealed the contribution of numerous non-coding RNAs (ncRNAs) to metastasis and invasion of gynecologic cancer by affecting specific cellular pathways. This review focuses on three types of gynecologic cancer (ovarian, endometrial, and cervical) and three kinds of ncRNAs (long non-coding RNAs, microRNAs, and circular RNAs). We summarize the detailed role of non-coding RNAs in the different pathways and molecular interactions involved in the invasion and metastasis of these cancers.
1 Introduction
Gynecologic cancer can affect various organs within the female reproductive system, including the uterus, cervix, vulva, ovary, and vagina. In 2020, there were 313,959 new cases of ovarian cancer, 417,367 new cases of endometrial cancer, and 604,127 new cases of cervical cancer reported worldwide, with recorded death numbers of 207,252, 97,370, and 341,831, respectively (1). Fortunately, the incidence of cervical cancer has decreased over the past three decades, thanks to routine screening, HPV vaccination, and the management of premalignant lesions. However, the incidence of ovarian and endometrial cancer has increased (2).
Metastasis is a multi-stage dynamic process that largely relies on the complicated interactions of tumors with the intrinsic host components and the microenvironment (3). Metastasis can only take place if the metastatic cancer cells can survive the physical insults encountered during their journey and avoid destruction by the host immune system. In order for the cells to multiply, migrate, and colonize distant tissues, they might need to lie dormant for lengthy stretches of time. Therefore, the attack by the host immune response must be avoided, and the immune cells can even be altered by the metastatic cancer cells (4). It is thus essential for the metastatic cancer cells to interact with host cells mediated by cytokines or extracellular vesicles and to undergo epithelial-to-mesenchymal transition (EMT). EMT allows the cancer cells to migrate and invade the surrounding tissues and to evade protective processes such as shear stress, immune susceptibility, and anoikis. These cells show more malignant characteristics at both the genetic and the phenotypical levels (5).
MicroRNAs (miRNAs) are RNA sequences that are roughly 22 nucleotides in length (6). miRNAs attach to the 3′UTR of targeted mRNAs by base pairing to block the post-transcriptional translation or trigger the degradation of the target mRNA. These miRNAs are capable of negatively regulating the expression of the target gene and can either inhibit or promote tumor metastasis, depending on the specific genes involved (7). lncRNA sequences are more than 200 nucleotides in length but do not code for any proteins. In addition, lncRNAs are capable of regulating gene expression in a variety of ways. These include direct binding or base complementation with the target gene to regulate its transcription and the indirect modulation of the downstream or upstream pathways related to the gene in question (8, 9). Although researchers have shown the contribution of some lncRNAs to tumor formation, further research is needed into the underlying mechanisms of how lncRNAs can affect metastasis (10). Circular RNAs (circRNAs) are more stable than linear RNAs and contain a linkage between the 5′ splice site in the downstream direction and the 3′ splice site in the upstream direction. The biogenesis of circRNAs involves lasso driving, intron cyclization, or intron pairing. Some researchers believe that circRNAs are a by-product of splicing errors and thus were primarily ignored in previous investigations. Nowadays, many circRNAs have been discovered, thanks to major improvements in sequencing technology (11, 12).
2 Metastasis and gynecological cancer
Oncogenesis is a complex process that involves multiple steps and the accumulation of several mutations that affect cell proliferation and equilibrium. Metastasis, which is the spread of cancer cells from the primary tumor to distant tissues and organs, is another complicated process that relies on the activation of several mechanisms. These mechanisms include angiogenesis, infiltration, embolization, survival in the bloodstream, arrest in organs, attachment to vessel walls, and extravasation (13). To initiate and control tumor progression and metastasis, cancer cells secrete cytokines, and regulatory immune cells play a crucial role in these processes. In response to cellular damage and stress, immune cells release cell signaling molecules that modify immune reactions, reducing cell injury and boosting cell development (14). However, cancer cells can bypass the immune system’s innate and adaptive defenses by generating antigens (15, 16). The tumor cells interact with the organ environment, known as the “soil and seeds hypotheses,” which is believed to cause metastasis (17). The cancer cells from the initial tumor are the seeds, and the metastatic site is the soil. Metastasis is the leading cause of mortality for more than 90% of cancer patients, including those with gynecological cancers. Gynecological cancers, such as ovarian and cervical cancer, are caused by genetic mutations that affect cell proliferation and equilibrium. These mutations are randomly produced by damage to DNA and lack or malfunction of DNA repair systems. The mechanisms involved in initiating and advancing metastasis in gynecological cancers include invasion, circulation, intravasation, extravasation, and colonization (Figure 1).
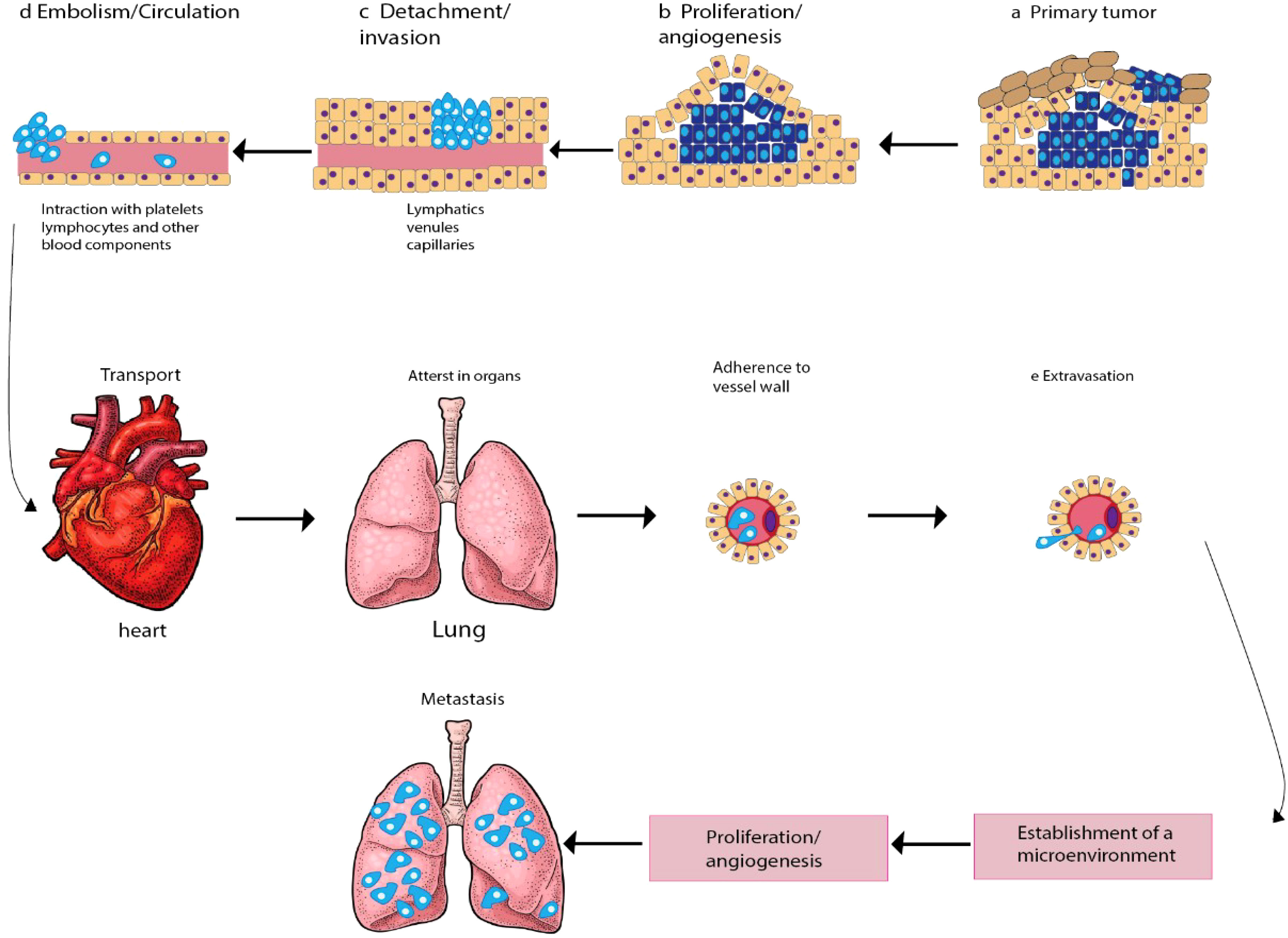
Figure 1 Schematic diagram depicting the main steps in the formation of a metastasis. The progression of cancer metastasis involves a series of selective steps that are influenced by interactions between metastatic cells and homeostatic factors. Failure of a tumor cell to complete any step effectively terminates the process. Consequently, the formation of clinically relevant metastases reflects the survival and growth of distinct subpopulations of cells that already exist within primary tumors. (A) The process begins with cellular transformation and tumor growth. (B) Extensive vascularization should occur if the tumor size increases. This is achieved through the synthesis and secretion of angiogenic factors, which establish a capillary network from the surrounding host tissue. (C) Some tumor cells migrate and invade the host stroma via several parallel mechanisms. Lymphatic channels offer little resistance to penetration by tumor cells and are the most common route for tumor cell entry into the circulation. (D) Subsequently, detachment and embolization of single tumor cells or aggregates occur; most circulating tumor cells are quickly destroyed. Once cancer cells survive in the circulation, they become trapped in the capillary beds of distant organs by adhering to either capillary endothelial cells or the subendothelial basement membrane. (E) Extravasation then occurs, likely through mechanisms similar to those during invasion. Proliferation within the organ parenchyma completes the metastatic process. To continue growing, the micrometastasis must develop a vascular network and evade destruction by host defenses. The cells can then invade blood vessels, enter the circulation, and create new metastases.
2.1 Invasion
Invasion is the process by which cancer cells break away from the primary tumor and invade surrounding tissue. Epigenetic factors induced by environmental stimulation, such as adhesive signals from extracellular matrix (ECM) components, aging, and circadian disruptions as well as cell–cell interactions, soluble signals, and the intratumoral microbiota, can all contribute to the activation of invasion and metastasis in gynecological cancers. Cancer cells can invade the surrounding tissue by secreting enzymes that break down the extracellular matrix, which is a network of proteins and fibers that provide structural support to tissues (18). In gynecological cancers, this can involve the invasion of nearby organs such as the ovaries, fallopian tubes, uterus, cervix, vulva, or vagina. According to in vivo and in vitro research, metastatic cancer cells move independently. In humans, however, seeding needs the coordinated activity of a group of tumor cells, which brings EMT into play (19, 20). EMT is a biological mechanism in which epithelial cells lose their properties and take on mesenchymal traits. Apical–basal polarity, cell–cell junctions, and epithelial markers are lost when epithelial cells undergo EMT, whereas a spindle-cell shape, cell motility, and mesenchymal markers are gained (21). Once the cancer cells have invaded the surrounding tissue, they can enter the bloodstream or lymphatic system.
2.2 Intravasation
Cancer cells are disseminated to organs through the vascular lumen, either actively or passively. Intravasation is the step that happens following the invasion. Intravasation is the process by which cancer cells enter the bloodstream or lymphatic system (22). In gynecological cancers, cancer cells can enter the lymphatic system through the lymphatic vessels that surround the reproductive organs or the bloodstream through the rich vascular supply of the reproductive organs. Once cancer cells have entered the circulation, they can travel to other parts of the body.
2.3 Circulation
During the circulation stage, cancer cells travel through the bloodstream or lymphatic system to distant sites and organs. Cancer cells may be subjected to mechanical and immune clearance during this stage, but some cancer cells can survive in the circulation by evading the immune system or by forming clusters called emboli that can block small blood vessels and protect the cells from shear stress and immune clearance.
2.4 Extravasation
Extravasation is the process by which cancer cells leave the circulation and invade a new tissue.
In gynecological cancers, cancer cells can extravasate into the ovaries, fallopian tubes, uterus, cervix, vulva, or vagina. The ability of cancer cells to extravasate depends on their interaction with the endothelial cells that line the blood vessels in the target organ and their ability to penetrate the extracellular matrix. Extravasation is a complicated process involving ligand–receptor interactions, chemokines, and non-tumor cells in the bloodstream. Integrins play a role in oncogenic growth factor receptor (GFR) signaling and GFR-dependent cancer cell motility and invasion, facilitating the anchorage-independent survival of circulating tumor cells (CTCs) and in governing the colonization process in metastatic sites. Chemokines and complement components can direct tumor cells to specific locations (23). When cancer cells are packed, they produce more IL-6 and IL-8, two immune chemicals that trigger biochemical pathways and aid in tumor migration (24, 25). Cancer cells may migrate alone or in groups. CTCs can extravasate and populate new habitats after being arrested at secondary locations or trapped in capillaries Integrins, once again, play an important role in defining the locations of extravasation and colonization by allowing CTCs to survive without anchoring (22, 23). Once cancer cells have extravasated, they face hostile environments that make life challenging. Some cells fall into dormancy as a response to the new stressful environment (18). The creation of the premetastatic niche, in which the tumor cells infiltrate and thrive, is triggered by various secreted tumor-derived substances and bone marrow-derived cells (26).
2.5 Colonization
Colonization is the final stage in metastasis, where cancer cells establish a new tumor in the new tissue. The ability of cancer cells to colonize a new tissue depends on a number of factors, including the ability of the cancer cells to adapt to the new environment, the presence of growth factors that can stimulate the growth of new blood vessels, and the ability of cancer cells to evade the immune system.
In gynecological cancers, such as ovarian and cervical cancer, several molecular variables are linked to metastasis, including HOX genes, PI3K/AKT/mTOR signaling pathway, EGFR, platelet-derived growth factor receptors, and vascular endothelial growth factor (VEGF) (27)—for instance, the ovulatory cycle-induced angiogenesis, the presence of COX-1, and the availability of growth factors offer an ideal environment for the implantation of glioma-initiating cells (GICs) in ovarian cancer (OC). Ovarian cancer commonly presents at advanced stages and can spread through both passive and hematogenous mechanisms (Figure 2) (28). Metastatic ovarian cancer (MOC) accounts for 2.3% to 23.7% of all malignant ovarian tumors that are generally transmitted from other organs. MOC most often arises from the gastrointestinal (GI) tract (71%), followed by the appendix (8%), breast (6%), and pancreas (4%), according to a recent research study in Japan. MOC differs from other gynecologic cancers. It has non-obvious symptoms in the early stages (abdominal mass and/or fullness is the most prevalent symptom) and no characteristic imaging findings (29). Compared to older female GIC patients, younger female GIC patients in the ovulatory period are more likely to develop MOC (30). The ovary’s ovulatory cycle, according to researchers, creates a perfect environment for GIC cells to survive and penetrate (31). When an oocyte is released to repair the surface of the ovary following ovulation, the epithelium of the ovary is disturbed by the buildup of steroid hormones. It is comparable to wound healing, which necessitates the formation of new blood vessels (32). According to other studies, the ovary has all of the VEGF-A isoforms, and both VEGFR-1 and VEGFR-2 are extensively expressed in ovarian capillaries (33). Angiopoietin-2 was expressed in the ovary, which is noteworthy (34). Furthermore, numerous factors such as oxygen saturation, age, and endocrine function impacted the expression of angiogenic peptides. The ovary contains gonadotropic hormones such as luteinizing hormone (LH) and follicle-stimulating hormone (FSH). LH and FSH control ovarian angiogenesis by raising the VEGF levels dose-dependently (35). Moreover, LeCouter et al. (2001) discovered the first tissue-specific angiogenic molecule in ovarian tissue, which was obtained from the endocrine gland (36). Other variables and ovarian angiogenesis increase GIC cell growth, seeding, invasion, and survival. COX enzymes have been shown to transfer to eicosanoids, which have been shown to promote GIC cell transformation and proliferation. COX is also linked to the existence of VEGF, which was previously explored. COX-1 expression was abundant in both normal and malignant ovarian tissue, while VEGF was abundant in the same areas. COX-1 seems to enhance neovascularization and cell proliferation, according to these data. GIC cells metastasizing to the ovary are also regulated by other growth factors such as epidermal growth factor, hepatocyte growth factor, and TGF. In conclusion, the ovulatory cycle-induced angiogenesis, the presence of COX-1, and the availability of growth factors offer an ideal environment for the implantation of GIC cells (37, 38). Cervical cancer development and metastasis are caused by genetic changes in multiple cell signaling systems that influence the choice of apoptosis or survival.
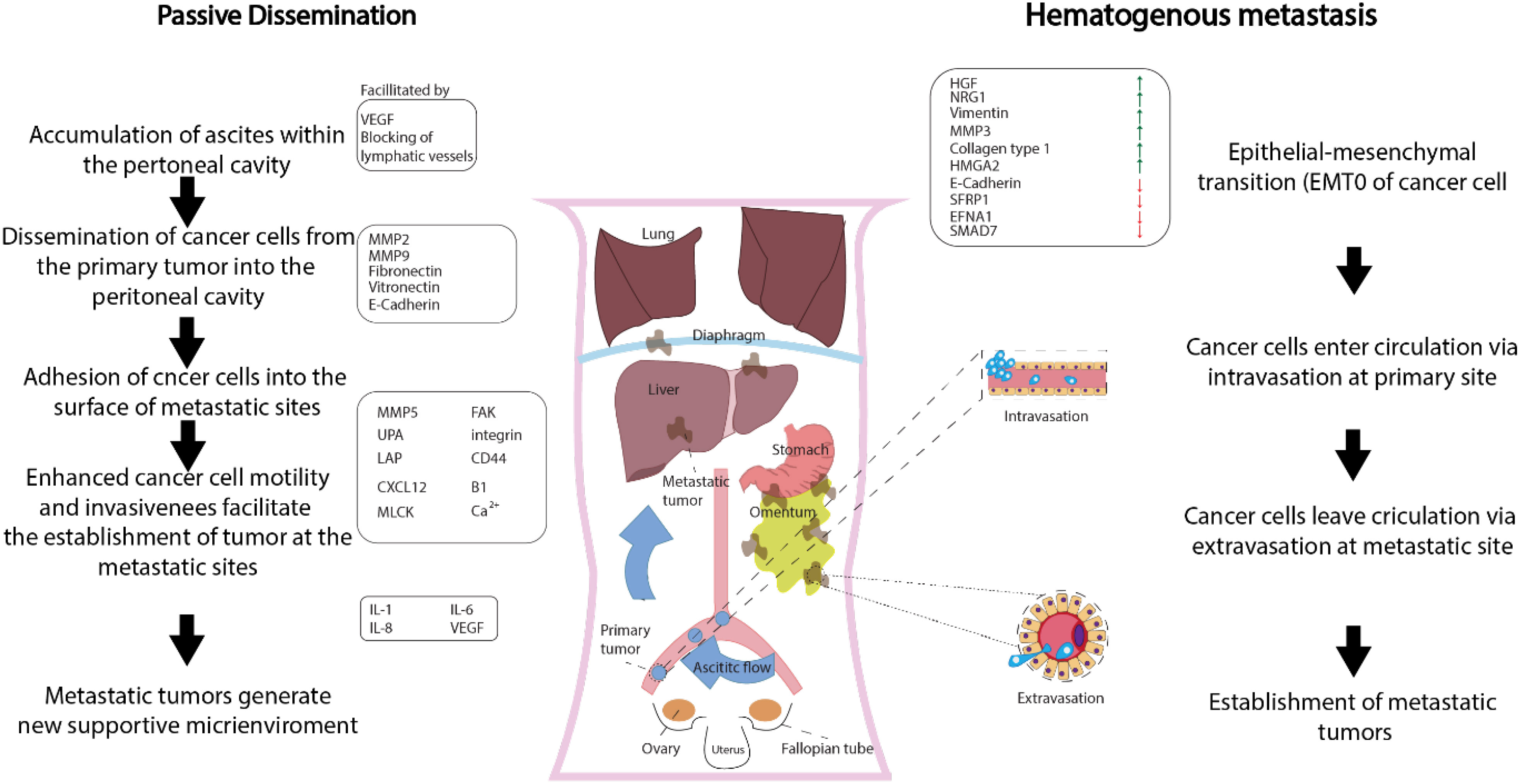
Figure 2 Metastasis of ovarian cancer on a molecular level (approved by the American Physiological Society).
In summary, understanding the mechanisms involved in tumor progression and metastasis is crucial for developing effective therapies for gynecological cancers. Targeting the molecular variables related to metastasis and blocking each of the steps involved in it may be effective strategies for the prevention and treatment of female metastatic cancers.
3 ncRNAs and metastasis in gynecological cancer
In gynecological cancers, ncRNAs have been implicated in regulating various biological processes associated with metastasis, such as invasion, angiogenesis, and immune evasion. In addition to their roles in regulating metastasis-associated processes, ncRNAs have also been shown to play important roles in regulating the tumor microenvironment. Emerging evidence suggests that dysregulation of ncRNAs is involved in many aspects of cancer, including tumor progression and metastasis.
3.1 miRNAs and metastasis in gynecological cancer
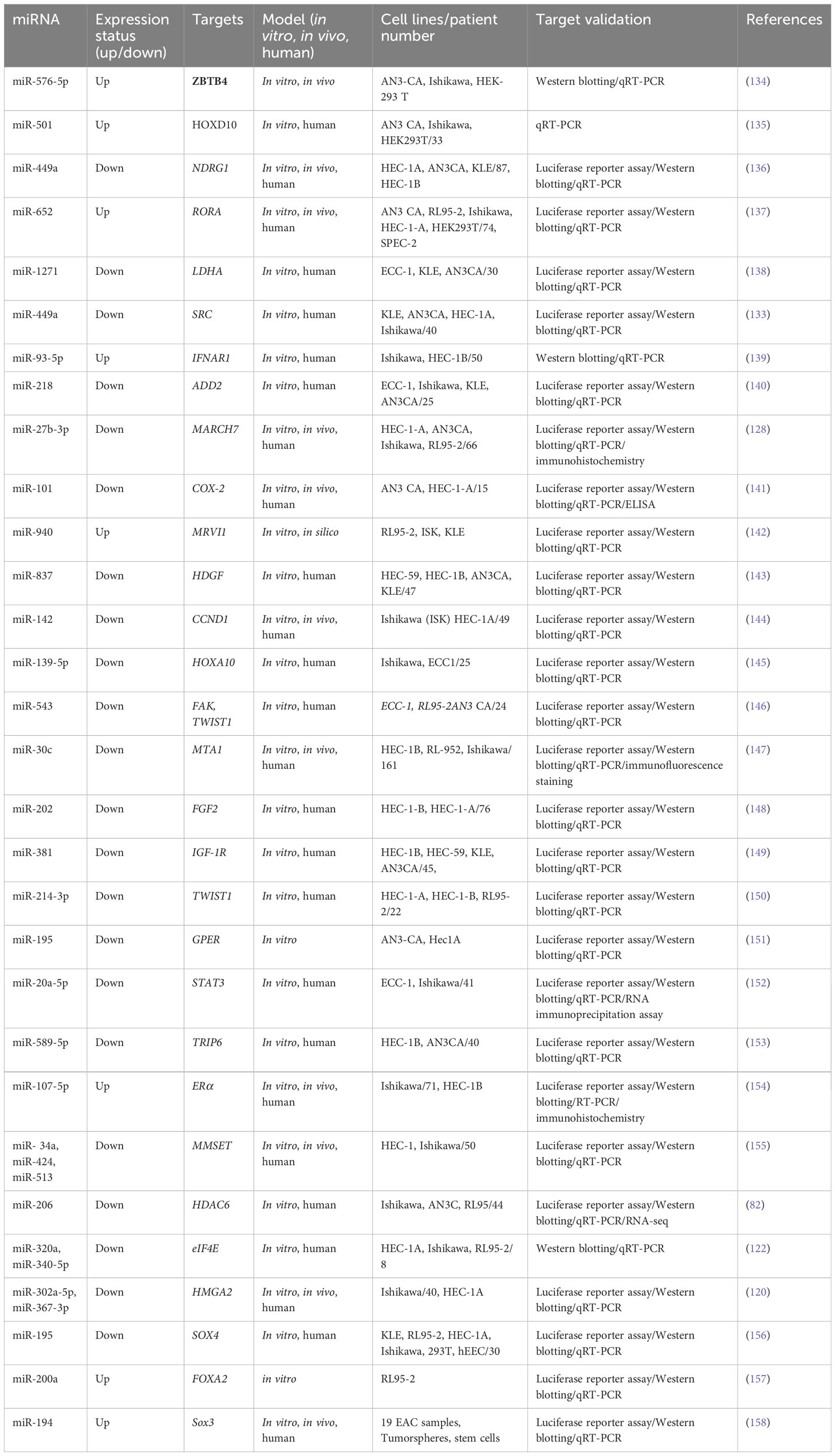
3.1.1 Metastasis-related miRNAs in ovarian cancer
OC has the 14th rank of cancer-attributed mortality among both sexes worldwide (1). Moreover, the 5-year survival of I–II stages varies from 75% to 92%, but around one-third of patients in Western countries are still diagnosed with advanced peritoneal dissemination and ascites (39). The development of a practical and sensitive approach for the early detection of ovarian cancer is required to reduce the high death rates. Unfortunately, the early stages of this disease are often not detected by recent diagnostic methods, such as CA125 serum levels, pelvic examination, or transvaginal ultrasound (40).
One approach to discovering diagnostic and prognostic biomarkers for ovarian cancer relies on the different levels of expression of certain miRNAs in plasma, ascites fluid, serum, serum exosomes, or tissue biopsies taken from ovarian cancer patients and healthy controls. One study of tissue miRNA expression profiles collected from subjects with ovarian cancer and healthy individuals showed distinct miRNA signature profiles between the two groups. All morphological histotypes of ovarian cancer tissue were included, showing typically elevated levels of miR-141 and miR-200a-c, which typically reduced the miR-125b, miR-199a, miR-140, and miR-145 levels. Furthermore, different miRNA patterns were found in ovarian cancer samples with different histopathological characteristics, i.e., serous, mucinous, and endometrioid as well as clear cell—for instance, miR-212 and miR-302b* were greatly elevated, whereas miR-222 was reduced in the endometrioid histotype compared to the serous histotype (41).
A study by Fu et al. (2016) demonstrated that miR-222-3p targets GNAI2 in epithelial ovarian cancer, leading to the suppression of tumor cell proliferation (42). However, in contrast, another study in endometrial cancer showed that miR-222-3p targets the estrogen receptor (ERα), leading to increased cell proliferation and tumor spread (43). Furthermore, miRNAs can have specific antagonistic activities in certain cancer stages or types (44). Further investigation is needed to fully understand the inhibitory impacts of miR-222-3p on cell migration in epithelial ovarian cancer.
The CCM family of proteins includes cerebral cavernous malformation 3 (CCM3), krev-interaction trapped 5 (KRIT5), and programmed cell death 10 (PDCD10) (45, 46). These three CCM family members (CCM2, PDCD10, and KRIT1) have been shown to have critical regulatory effects on endothelial cell–cell interactions and vascular equilibrium (47). In addition, the interaction between PDCD10 and MST4 stabilizes each of them so that PDCD10 can stimulate MST4-dependent cell proliferation and migration (48). Moreover, PDCD10 and germinal center kinase III (GCKIII) can interact with each other, affecting the serine/threonine-protein kinases STK25 and STK24 (49). In a mechanistic study, Fan et al. (2020) investigated the regulatory function of miR-222-3p in EOC, which could help improve the current anti-metastasis therapy. The target genes of miR-222 were predicted using four separate prediction databases of miRNA targets. Moreover, binding between 3′-UTR of the PDCD10 mRNA and miR-222-3p was confirmed using a luciferase assay. In the study, the authors also applied transwell migration and scratch wound healing assays as well as a xenograft mouse model to explore the biological activities of miR-222-3p and PDCD10. They predicted the ability of transcription factor SNAI2 to alter the expression of miR-222-3p using UCSC, JASPAR, and ENCODE public databases. The supposed SNAI2 binding sites for miR-222-3p were confirmed using a luciferase reporter assay. In addition, the researchers investigated SNAI2 binding to the miR-222-3p promoter using chromatin immunoprecipitation. They discovered that SNAI2 downregulated miR-222-3p in EOC tissues and cells, and this suppressed tumor formation. The bioinformatics database revealed that PDCD10 negatively correlated to miR-222-3p, both in vivo and in vitro. They found that miR-222-3p rapidly binds to the 3′-UTR of PDCD10, inhibiting its translation and EOC cell migration in vitro and inhibiting EOC xenograft tumor spread in vivo. The over-expression of PDCD10 downregulated E-cadherin, but upregulated vimentin, and stimulated the EMT and β-catenin/Wnt-mediated cell migration, all of which ultimately tended to increase metastasis (50).
Many miRNAs have been shown to contribute to OC development and progression. One of these is miRNA-6089, which has recently been found to be involved in OC development. Moreover, over-expression of miR-6089 inhibited the rapid growth of the ovarian cancer cells and infiltration and reduced metastasis in vivo, according to a study conducted by Liu and colleagues (2020). Recent studies showed that miR-6089 inhibited Wnt/β-catenin signaling and the associated EMT and reduced the expression of c-Jun and cell-cycle mediators via direct targeting of MYH9. The over-expression of MYH9 led to the upregulation of Wnt/β-catenin and EMT, c-Jun, and cell cycle mediators, thus abrogating the inhibitory effect of miR-6089 upregulation on ovarian cancer. c-Jun is one of the transcription factors which is activated by MYH9 via the Wnt/β-catenin pathway, suppressing miR-6089 production. In ovarian cancer, the miR-6089/MYH9/β-catenin/c-Jun axis acts as a negative feedback loop. miR-6089 expression was shown to be inversely associated with MYH9 expression in clinical specimens. Therefore, miR-6089 acts as one of the tumor-suppressor miRNAs in ovarian carcinogenesis and cancer development (51).
miR-489 is a miRNA that has been shown to play a role in tumor biology (52). In glioma cells, miR-489 was found to trigger apoptosis and decrease cell proliferation by modulating the SPIN1-mediated phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) pathway (53). In ovarian cancer, miR-489 has also been shown to downregulate Akt3, which enhances apoptosis, reduces cell proliferation, and overcomes cisplatin resistance. A study by Wu et al. (2014) demonstrated the effectiveness of miR-489 in enhancing the sensitivity of ovarian cancer cells to cisplatin (43). In human tissues, X-linked inhibitor of apoptosis protein (XIAP) is a powerful suppressor of apoptosis (54), which has recently been confirmed to be a tumor suppressor (55). The contributions of miR-489 and XIAP to OC progression, invasion, and metastasis were recently investigated (56). Expressing miR-489 in OC tissue samples and cell line has been confirmed with the use of qRT-PCR. Moreover, the miR-489 levels of OC tissues and cells have been significantly lower than those in normal controls and were linked with malignant clinical pathologic characteristics and a poor prognosis in OC patients. miR-489 was found to inhibit OC cell viability, invasion, and migration in functional tests. XIAP was shown to be a miR-489 target, partly responsible for its effects in OC. miR-489 also suppressed OC development via modulating the PI3K/AKT pathways and the EMT. miR-489 reduced OC progression by directly binding to XIAP mRNA and modulation of the PI3K/Akt and EMT signaling pathways, revealing that it is possibly used as a biomarker for OC prognosis and therapy in the future (56).
Emerging evidence suggests that miR-338-3p plays a role in the initiation and progression of several human cancers, including rectal, liver, gastric, lung, and neuroblastoma. In these malignancies, miR-338-3p has been shown to act as a tumor suppressor, inhibiting invasion and the migration of cancer cells (57). The role of miR-338-3p in OC has been studied in only a limited number of reports. One study found that miR-338-3p inhibits OC cell growth and metabolism, suggesting a potential tumor-suppressive role for this miRNA. Another study showed that miR-338-3p can inhibit the development of ovarian epithelial cancer by targeting Runx2, a protein involved in the regulation of cell proliferation and differentiation (58). In epithelial ovarian cancer tissues, researchers showed that miR-338-3p reduced and was negatively associated with the MET transcriptional regulator metastasis-associated in colon cancer protein 1 (MACC1) (59). However, additional reports regarding the function of miR-338-3p in OC should be required. Zhang et al. (2019) designed a study to investigate the contribution of miR-338-3p to the proliferation of the OC cells and metastasis, along with the associated molecular mechanisms (60). The researchers used a multi-biomedical database query and a “‘KEGG pathway enrichment test to identify the potential target genes as well as the downstream pathways affected by miR-338-3p. Colony formation, MTT, transwell, and Matrigel migration assays as well as a xenograft mouse model, were used to measure proliferation, migration, and invasion after lentiviral vectors were used to over-express miR-338-3p in OVCAR-8 and OVCAR-3 ovarian cancer cells. Western blotting was performed to measure MACC1 (a miR-338-3p binding target gene) and MET and the downstream signaling pathways. A search of biomedical databases showed that miR-338-3p could affect MET, the MEK/ERK pathway, and downstream Wnt/β-catenin along with the MACC1 gene. Replacement of miR-338-3p might inhibit the rapid growth of the OC cells, migration, and invasion and reduce xenograft tumor development and metastasis. Over-expression of MACC1 and Met promoted MEK/ERK activity, proliferation, EMT, and Wnt/β-catenin, all of which could be reduced if miR-338-3p was restored. In conclusion, miR-338-3p suppressed OC metastasis and rapid growth, perhaps via suppressing EMT caused by Met, Wnt/β-catenin, MEK/ERK signaling, and MACC1 (60).
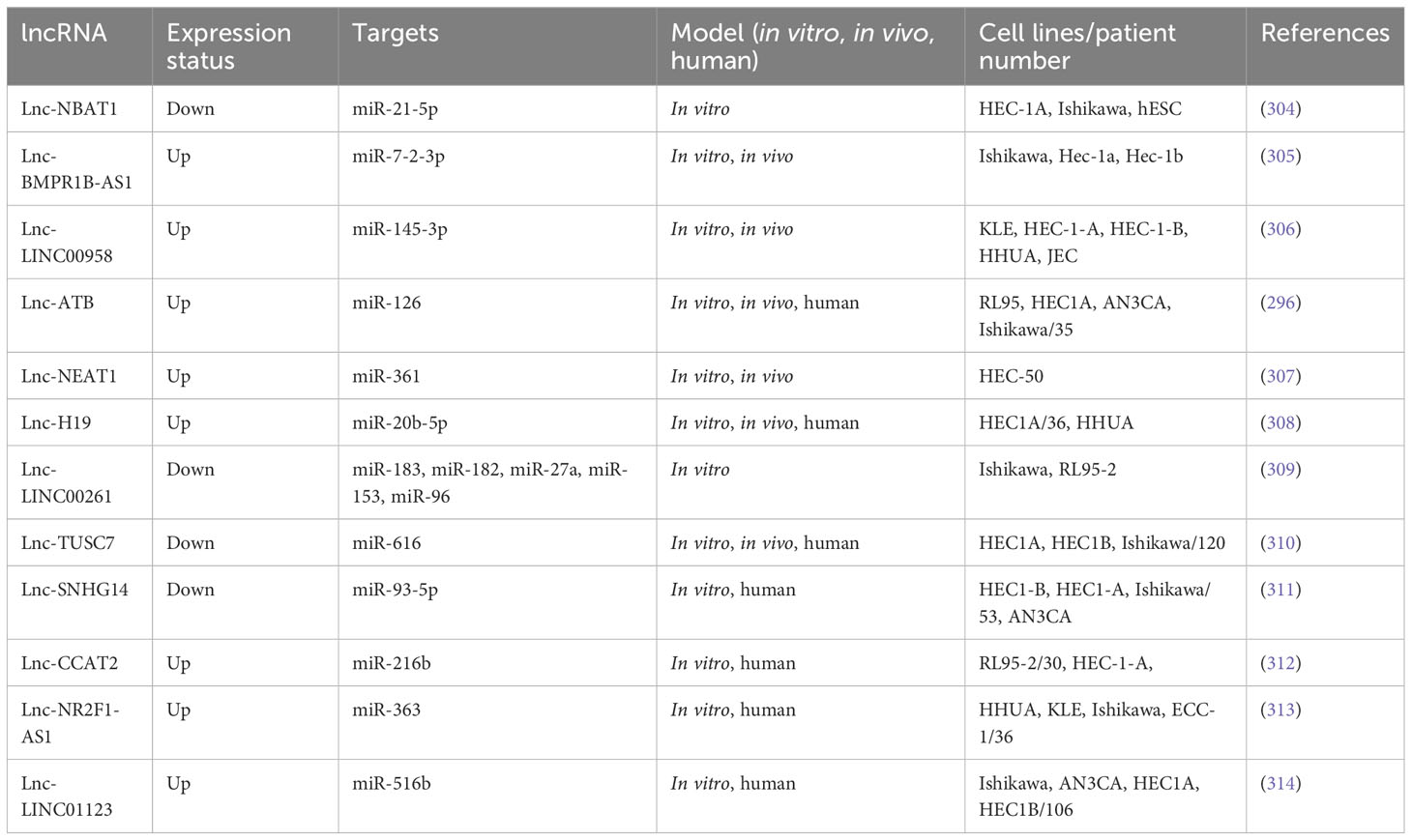
The dysregulation of miR-936 levels has been linked to NSCLC and glioma progression, but the activity of miR-936 has rarely been discussed in EOC. miR-936 upregulation reduced proliferation, caused cell cycle arrest, and reduced invasion in NSCLC tissues and cell lines (61). In glioma tissue and cell lines, expressing miR-936 was similarly reduced. Cases with a low expression level of miR-936 demonstrated a worse prognosis than those with higher levels of miR-936 expression. Li et al. (2019) designed an experiment to study miR-936 expression in EOC and its mechanism of action. Researchers employed RT-qPCR for measuring miR-936 expression in EOC. Flow cytometry, CCK-8 assay, migration, invasion assays, and a xenograft nude mouse model were employed to assess apoptosis, migration, invasion, rapid growth in vitro, and tumor development in vivo. The relationship of miR-936 with FGF2, a highly expressed prototypical growth factor in numerous cancers, was investigated using bioinformatics, RT-qPCR, Western blotting, and luciferase reporter assays. EOC cells and tissues showed dramatically lower expression levels of miR-936. Furthermore, in EOC patients, lower miR-936 expression has shown a correlation to the FIGO stage and the size of the tumors as well as the presence of lymphatic metastasis. The ectopic expression of miR-936 inhibited migration, proliferation or rapid growth, and invasion, increased cell apoptosis in vitro, and reduced tumor development in vivo. Moreover, in EOC cells, the FGF2 gene has also been found to be directly targeted by miR-936. FGF2 expression was elevated in the EOC tissues, which was negatively correlated to the miR-936 expression. In addition, FGF2 silencing in EOC cells led to similar results to miR-936 over-expression. In EOC cells, the restored levels of FGF2 reversed the inhibitory effects of miR-936 and controlled FGF2 to inhibit the PI3K/Akt signaling pathway in vitro and in vivo. Overall, their findings demonstrated thatmiR-936, at least in part, suppresses the metastatic behavior of EOC cells in vitro and in vivo via affecting the FGF2-mediated regulation of PI3K/Akt and could act as a therapeutic target. Table 1 shows the contribution of some miRNAs to OC metastasis (93).
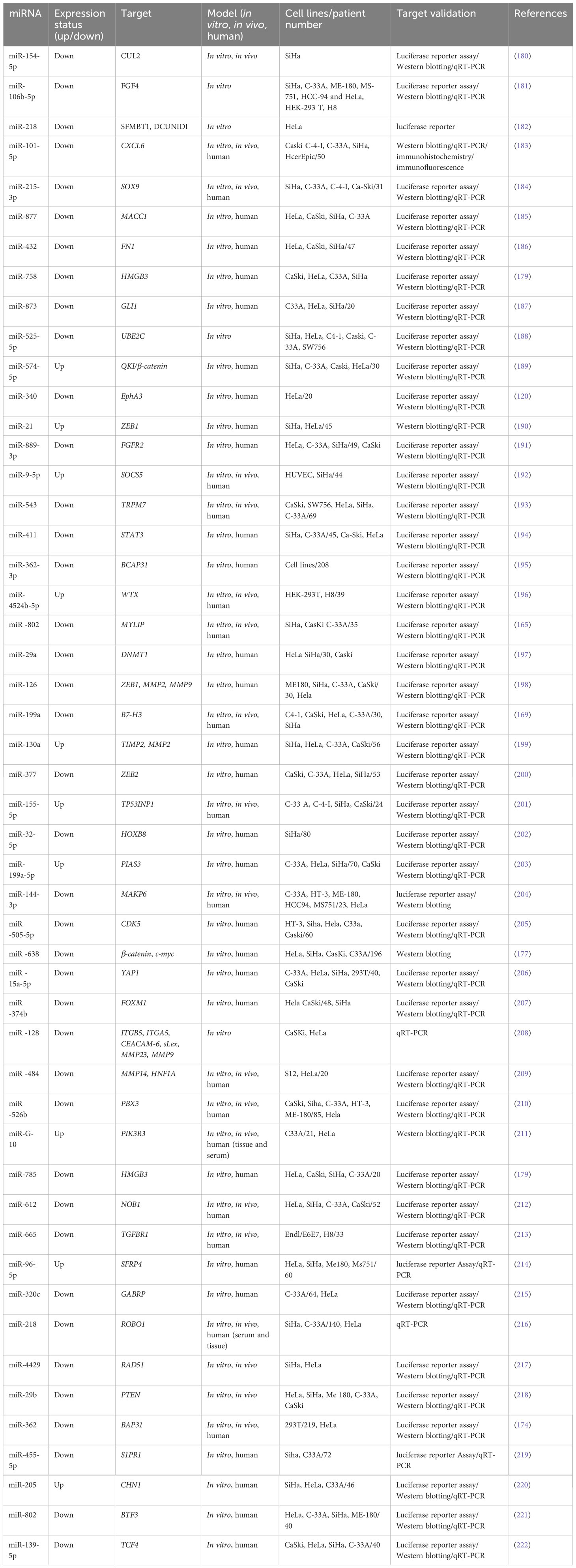
3.1.2 Metastasis-related miRNAs in endometrial cancer
Endometrial cancer (EC) has the 19th rank of cancer-attributed mortality among both sexes worldwide (1). Endometrial cancer is categorized into two subtypes. Type I tumors are frequently preceded by endometrial hyperplasia and are usually endometrioid adenocarcinomas associated with unopposed estrogen stimulation and extreme obesity (113). Type II tumors arise in atrophic endometrium as primarily serous carcinomas, which are estrogen-independent and less differentiated, with a lower survival rate (113). Fortunately, most endometrial cancer cases are type I endometrioid, which have a better prognosis (114). This is primarily due to the fact that women with vaginal bleeding tend to seek treatment earlier, so their disease is diagnosed at an earlier stage (115). The most recent findings indicate a 5-year survival rate of 48.7% for FIGO stage III and 28.2% for FIGO stage IV disease (116).
Lower levels of miR-206A have been shown in a variety of malignancies, including rhabdomyosarcoma and lung and breast cancer. However, further investigations are needed to understand the role of miR-206 in EC (117). Researchers categorized histone deacetylase (HDAC) enzymes into four categories: class I (HDAC1, HDAC2, HDAC3, and HDAC8), class II (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10), class III (SIRT1– SIRT17), and class IV (HDAC11). HDAC enzymes eliminate the acetyl groups (O=C–CH3) from the N-acetyl lysine amino acids in histone proteins to allow tighter wrapping of genomic DNA and modulate gene expression (118). HDAC6 is a unique HDAC, predominantly functioning in the cytoplasm, unlike other HDAC types. HDAC6 expression has been frequently linked to oncogene mutations and the progression of cancer, including ovarian and breast tumors (119). Zheng et al. (2020) analyzed the role of HDAC6 in EC diagnosis and treatment. Bioinformatics and dual-luciferase experiments showed that miR-206 could directly target HDAC6 mRNA. They found that HDAC6 exerted an opposite effect compared to miR-206 by promoting EC cell metastasis, invasion, and proliferation, with colony formation, CCK-8, and scratch wound healing as well as transwell assays. According to rescue tests, HDAC6 could reverse the effect of miR-206, and a bioinformatics analysis of gene expression validated the connection between the two genes. By measuring the levels of molecules such as PTEN, p-mTOR, and p-AKT, they suggested that miR-206 targets HDAC6 to inhibit EC development through the PTEN/AKT/mTOR pathway. miR-206 downregulation and HDAC6 upregulation in EC were poor prognostic indicators in EC patients (82).
miR-340 is another miRNA involved in several tumors. miR-340 is lower in cervical cancer, which inhibits the spread of cervical cancer by targeting ephrin-A-receptor 3 (120). miR-340-5p prevented breast cancer cells from developing drug resistance and inhibited proliferation. It also reduced the expression of leucine-rich repeat consisting of the G-protein coupled receptor 5 (LGR5) via the Wnt/β-catenin pathway, thus enhancing apoptosis (121). The eukaryotic translation initiation factor 4E (eIF4E) contributes to the regulation of protein production. Zhang et al. (2020) found an association between high eIF4E expression and poor prognosis in patients with high-pathological-grade EC using the Oncomine database microarray data. When comparing EC tissues to neighboring normal tissues, eIF4E expression has been shown to be greater in EC tissues. Furthermore, the miR-320a and miR-340-5p levels of expression have been higher in neighboring normal tissues in comparison with the EC tissues, suggesting that these two miRNAs were suppressor genes in EC. Both miR-340-5p and miR-320a bound to the 3′UTR of eIF4E mRNA and reduced the levels of eIF4E and phosphorylated eIF4E (p-eIF4E) in EC cells. Furthermore, HEC-1A cell invasion and migration were substantially reduced by the over-expression of either miR-320a or miR-340 5p. When miR-320a or miR-340-5p were transfected into cells, both eIF4E and p-eIF4E were downregulated, leading to lower expression levels of MMP3 and MMP9 and inhibition of EC invasion and metastasis. Furthermore, miR-320a and miR-340-5p upregulation inhibited the ability of TGF-β1 to trigger the phosphorylation of eIF4E. The TGF-β1-mediated EMT was likewise suppressed by these two miRNAs. To conclude, eIF4e has been greater in the EC tissue in comparison with adjoining normal tissues, and miR-340-5p and miR-320a were over-expressed in EC. Following the in vitro upregulation of the miR-340-5p or miR-320a, the migratory capacities of EC cells were reduced by inhibiting MMP3 and MMP9, and the TGF-β1-mediated EMT was blocked by p-eIF4E (122).
The membrane associated RING-CH (MARCH) protein family, which contains 11 members, is itself a part of the RING finger E3 Ubiquitin Ligase protein family. MARCH7, commonly referred to as axotrophin, has been shown to affect proliferation, migration, invasion, immunological tolerance, the actin cytoskeleton, autophagy, and neuronal development in both normal cells and cancer cells (123). MARCH7 was upregulated in developing rat spermatides during spermatogenesis, thus controlling the head and tail structural and functional properties (124). In mice, MARCH7 knock-down reduced the invasion and proliferation as well as migration of OC cells and prevented OC development (123). Research has shown that MARCH7, a protein that belongs to the MARCH family of E3 ubiquitin ligases, is involved in regulating cell and tissue growth and differentiation. Specifically, MARCH7 has been found to be expressed at higher-than-normal levels in stem cells, precursor cells, cancer cells, and certain other cells and tissues (125). A wide variety of transcription factors (TFs) have been found to be involved in the EMT, including Snail, Zeb, and Twist. These TFs, in turn, affect several tyrosine kinase receptor signaling pathways, including Hedgehog, β-catenin, TGF-β, STAT3, Notch, Wnt, and Nanog (126). In HUVECs, miR-27b-3p not only suppressed cell proliferation and migration via Smad7-mediated modification of TGF-β but also sensitized breast cancer cells to several anti-cancer treatments both in vivo and in vitro, suggesting the probable involvement of miR-27b-3p in cancer biology (127).
The involvement of MARCH7 in EC was investigated by Liu et al. (2019) (128). Moreover, the expression levels of MARCH7, Vimentin, Snail, and E-cadherin in the cell lines of EC and clinical tissue samples were investigated using Western blotting, immunohistochemistry, and quantitative polymerase chain reaction. The researchers employed a transwell assay and a xenograft tumor model to evaluate the involvement of MARCH7 in maintaining the malignant phenotype of EC cells. To test if MARCH7 is one of the direct targets of miR-27b-3p, the researchers employed a dual-luciferase reporter assay. MARCH7 expression in EC tissues was found to be higher compared to that in normal endometrial tissues. Moreover, the level of Vimentin and Snail, clinical stage, and histological grade were all positively correlated with MARCH7 levels, whereas E-cadherin levels were negatively correlated. Silencing of MARCH7 in vivo and in vitro reduced EC cell invasion and metastasis. By contrast, when MARCH7 was over-expressed, the opposite effect was found. MARCH7 increased EC cell invasion and metastasis by the Snail-mediated pathway. In addition, MARCH7 has been shown as a direct target of miR-27b-3p, so miR-27b-3p reduced the tumor-promoting impact of MARCH7. The above-mentioned findings suggest that MARCH7 is a tumor promoter factor, which could be a target in future EC therapy. The miR-27b-3p/MARCH7 axis interacts with the Snail-mediated pathway to control EC cell invasion and metastasis (128).
Another study has shown that the steroid receptor coactivator family (SRC-2, SRC-3, and SRC-1) was discovered to regulate the transcription of estrogen and progesterone receptors as well as other nuclear receptors (NRs) (129). SRC triggers a cascade of downstream signaling pathways, like PI3K/Akt pathways and MAPK/ERK, and regulates numerous cellular processes, particularly migration. SRC has been identified to be an important oncoprotein in many cancer types due to its strong regulation of NRs. Researchers have found the over-expression of SRC in several tumor types, such as breast cancer (130). In EC, SRC expression has a correlation to the clinical stage and unfavorable prognosis as well as depth of tumor invasion into normal tissue (131, 132). Hu et al. (2019) reported lower levels of miR-449a in advanced endometrial cancer cells. Furthermore, the AN3CA and KLE EC cell lines exhibited a weaker tendency to migrate and invade when miR-449a was over-expressed. SRC mRNA would be one of the direct targets of miR-449a, as shown by luciferase reporter assays. SRC expression has been greater in advanced EC tissues that had spread to distant sites. miR-449a could downregulate SRC to inhibit metastasis and reduce activating Akt and ERK1/2 pathways in EC cells (133). Table 2 shows the contribution of some miRNAs to endometrial cancer metastasis.
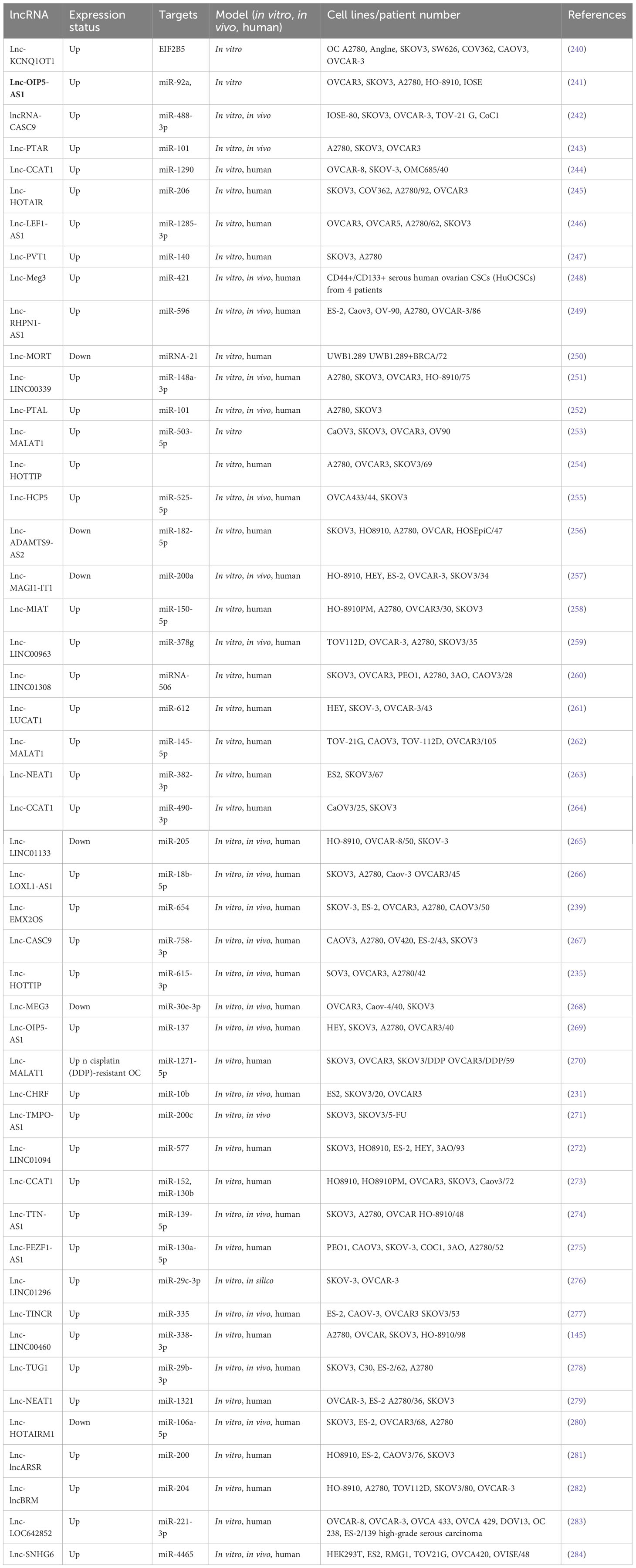
3.1.3 Metastasis-related miRNAs in cervical cancer
Cervical cancer (CC) is the fourth leading cause of death attributed to cancer among female patients worldwide (1). Long-term infections with higher-risk strains of human papillomavirus (HPV), like HPV-18 and HPV-16, account for the majority of CC cases (159). However, since some metastatic CC patients were found not to have had any HPV infection, it has been speculated that some unknown factors may be involved in the onset and progression of CC (160, 161).
Epithelial ovarian cancer, prostate cancer, and gastric cancer have all been found to be inhibited by miR-802 acting as a tumor suppressor (86). miRNA-802 can modulate serine/arginine-rich splicing factor 1 (SRSF1) to inhibit cervical carcinoma cell proliferation and promote cell death (162). The cytoskeletal protein cluster myosin regulatory light chain interacting protein (MYLIP) participates in cell migration (163). MYLIP contributes to cell motility, preservation of cellular morphology, remodeling of cytoskeletal proteins, and the adherence of cells to the ECM via interaction with cell membrane proteins (164). Ni et al. (2021) investigated the potential role of miR-802 in CC growth, invasion, and migration. The researchers used qRT-PCR to measure the expression levels of miR-802 and MYLIP in CC cells and tissues. They also employed a range of assays, including the CCK-8 assay, transwell invasion assay, scratch wound healing assay, and colony formation assay, to investigate the effects of miR-802 on CC cell proliferation and metastasis. In addition, an in vivo mouse xenograft model was used to examine the impact of miR-802 on CC development, and Western blotting and IHC were used to determine the MYLIP expression levels. The study found that the miR-802 levels were significantly lower in CC cells and tissues compared to normal cells and tissues. Higher levels of miR-802 were associated with reduced aggressiveness and slower growth of CC cells. The researchers also identified MYLIP as a direct target of miR-802 and found that it was over-expressed in CC. miR-802 could no longer suppress cervical cancer cell metastasis and proliferation when MYLIP was over-expressed. miR-802 inhibited the tumor growth of cervix in vivo, which also lowered MYLIP. In conclusion, miR-802 targets MYLIP for suppressing CC cell proliferation and metastasis (165).
B7-H3 is a B7 protein family member, which was found to be significantly expressed in tumors such as colon cancer (166, 167) while having minimal (if any at all) expression in most normal cells and tissues. Moreover, miR-199a has been found to play various roles in several cancers, depending on the kind of cancer. miR-199a was substantially lower in breast cancer and CC, where it targeted B7-H3 to modulate cancer development (168). Yang et al. (2020) demonstrated a reduction of miRNA-199a in the tissues of cervical cancer, while B7-H3 was considerably over-expressed compared to the surrounding normal tissue, as shown by qRT-PCR. They also found that miRNA-199a was lower in the cell lines of CC in comparison with the immortalized normal cells. Moreover, B7-H3 has been shown to be one of the targets of miRNA-199a in CC. The bioinformatics analysis results introduced 3′UTR of B7-H3 as one of the direct miR-199a targets, which was consistent with the results acquired from a luciferase reporter assay. Furthermore, the 3′-UTR of B7-H3 has been directly targeted by miRNA-199a; however, the exact signaling mechanisms that contribute to controlling B7-H3 expression have yet to be elucidated. A series of studies were carried out to see if the inhibitory action of miRNA-199a has been mediated by B7-H3. Over-expression of miRNA-199a repressed the proliferation and invasion as well as migration of cancer cells via binding directly to B7-H3. Cervical cancer metastasis was found to be dependent on the EMT. miRNA-199a suppressed tumor development in cervical cancer via targeting B7-H3, according to Western blotting and qRT-PCR. They also showed that miRNA-199a affected the Akt/mTOR signaling pathway via B7-H3 targeting and that over-expression of miRNA-199a suppressed tumor development in vivo. Their results could lay the groundwork for the development of future targeted prevention and treatment strategies for cervical cancer (169).
In a study conducted by Dang et al. (2018), B-cell receptor-associated protein 31 (BAP31) was found to be over-expressed in CC and to play a role in promoting tumor growth and progression. BAP31 is a cancer/testis antigen that is normally highly expressed in the testis and has been implicated in the development of various cancers. Additionally, BAP31 expression had a correlation to the CC clinical stage and stimulated the proliferation of the CC cells in vitro. As expected, the inhibition of BAP31 suppressed CC progression in vivo (170). Several cancers have been found to be suppressed by miR-362, which was downregulated in CC (171). miR-362 directly inhibited the expression of E2F1, USF2, and PTPN1, causing cell cycle arrest in colon cancer (172). miR-362 may also inhibit breast cancer progression by inhibiting the expression of p130 Crk-associated substrate (CAS) (173). Yang et al. (2021) discovered that miR-362 was negatively correlated with clinical stage in CC patients and was a major regulator of BAP31 expression. miR-362 over-expression reduced CC cell growth in vitro and increased apoptosis. Additionally, in a xenograft nude mouse model of CC, miR-362 decreased the tumor size and increased the mouse survival time. BAP31 binds to the spectrin isoform SPTBN1 to form a complex that modulates tumor development via the miR-362-regulated Smad 2/3 pathway. They showed that miR-362 was an anticancer, anti-proliferation, and pro-apoptotic miRNA in cervical cancer cells, which regulated the BAP31 and TGF-β/Smad pathways. Therefore, increasing the expression of miR-362 could be a possible cervical cancer treatment (174).
miR-758 over-expression has been observed in glioma and non-small lung cancer as well as hepatocellular carcinoma (175). miR-758 could act as a tumor inhibitor and prevent CC metastasis (176). miR-758 can also target matrix extracellular phosphoglycoprotein (MEPE) and inhibit infiltration and invasion in CC tissues (176). The high-mobility group box family, including HMGB1, HMGB2, HMGB3, and HMGB4, contributes to the progression of multiple cancers (177). In several cancers, including CC, the Wnt/β-catenin signaling pathway promotes cancer development (178). In colorectal cancer, HMGB3 was found to modulate the Wnt/β-catenin signaling pathway (177). Song et al. (2019) analyzed the effects of miR-758 on invasion, migration, and rapid growth in the CC cells. They used qPCR to show that miR-758 is considerably lower in CC tissues and the cell lines in comparison to normal controls. miR-758 over-expression significantly reduced viability, invasion, migration, and rapid growth, as shown by CCK-8, transwell, and colony formation assays. miR-758 inhibitors, on the other hand, increased these parameters. They showed that miR-758 directly targeted HMGB3 and that HMGB3 over-expression may counteract the impact of a miR-758 mimic on the viability, rapid growth, and invasion as well as migration of HeLa cells. miR-758 reduced HMGB3 expression that affected the Wnt/β-catenin signaling pathway and can play a part in new CC treatment strategies (179). The associations of some miRNAs to cervical cancer metastasis are listed in Table 3.
3.2 lncRNAs and metastasis in gynecological cancer
EMT is known as the key process responsible for the metastasis of different malignancies, which facilitates the transportation of malignant cells to distant areas (223). A number of intracellular signaling pathways have been identified to be involved in the induction of EMT. These signaling pathways become activated when the ligands from the stroma bind to their receptors on malignant cells. The bulk of evidence has existed in support of the fact that TGF-β/SMAD, Notch, PI3K/Akt, Wnt/β-catenin, MEK/ERK, and JAK/STAT signaling pathways have a mandatory role in inducing EMT-activating TF expression, in particular SNAIL, ZEB, and TWIST, which were shown to be able to activate and prohibit the expression of mesenchymal state-associated genes and epithelial state-associated genes, respectively (224). Recent shreds of evidence have demonstrated that EMT can be moderated by lncRNAs throughout the tumor metastasis process via regulating major molecules of a number of cellular and intracellular signaling pathways (225, 226) (Figure 3).
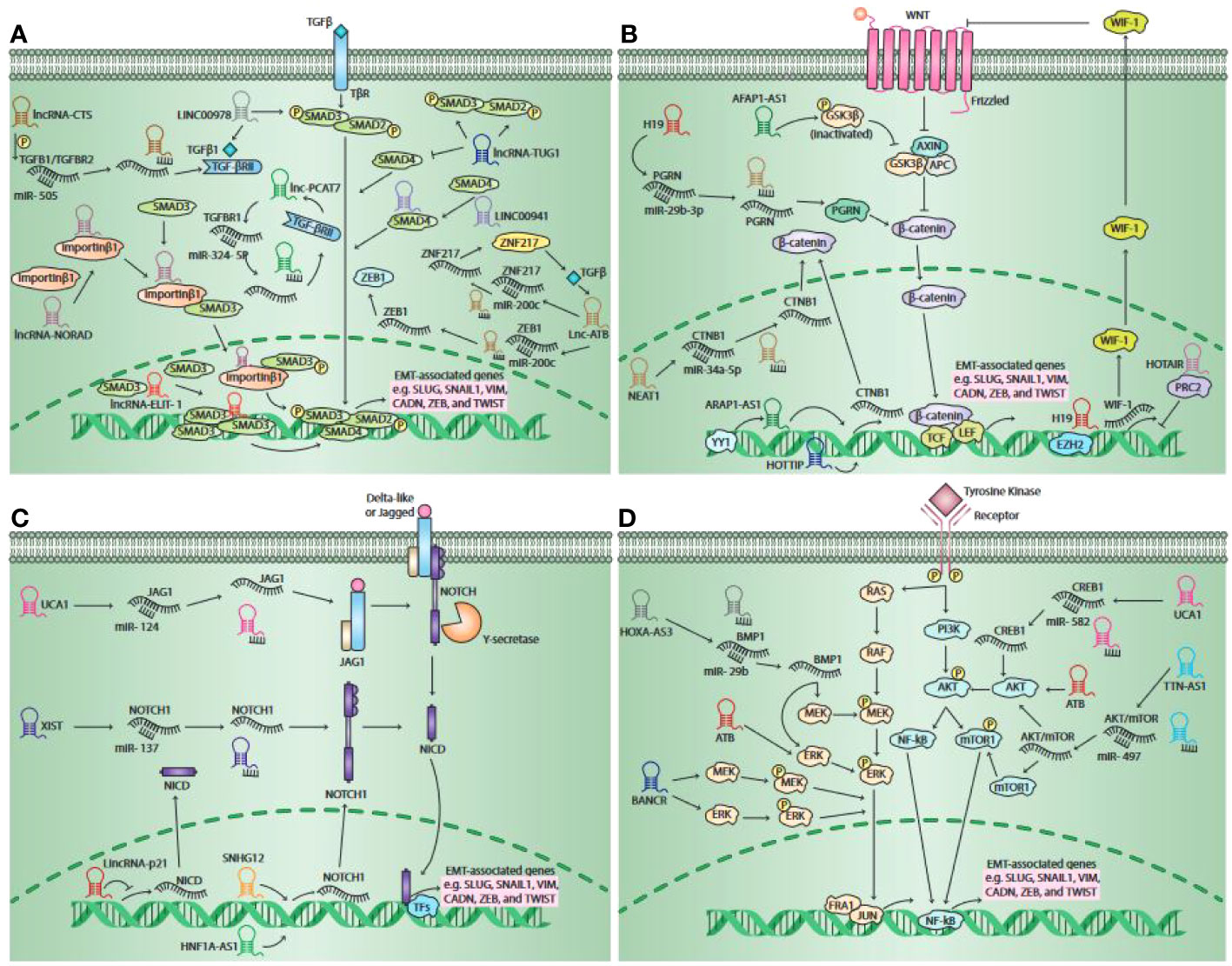
Figure 3 Schematic outline of the lncRNAs involved in pathways responsible for the activation of epithelial-to-mesenchymal transition (EMT). It has been unveiled that lncRNAs moderate EMT primarily via four main pathways, such as the Wnt signaling pathway, the TGF-β pathway, the Notch pathway, and the Mitogenic Growth Factor Signaling pathway. The activation of the TGF-β pathway occurs when canonical TGF-β ligands bind to their receptors, contributing to both SMAD2 and SMAD3 phosphorylation. When they become phosphorylated, they form a complex by binding to SMAD4. Thereafter, the complex travels to the nucleus and serves as a transcription factors to over-express EMT-related gene expression, including SNAIL1, CADN, SLUG, etc. lncRNAs are able to act as a signal molecule. LINC00978 mediates TFG-β/SMAD signaling transduction through activating SMAD2. It has been shown that lncRNA-TUG1 has the potential to enhance the phosphorylation of SMAD2 as well as SMAD3, whereas reducing the SMAD4 expression. LINC00941 was shown to be potentially activating TGF-β signaling via binding to SMAD4. lncRNAs were shown to have the potential to serve as ceRNA for some specific miRNAs. lncRNA-CTS over-expresses TGF-β1 and TGF-βRII expression via binding to miR-505, lncRNA-ATB over-expresses ZNF217 and ZEB1e expression through binding to miR-200c, and lncRNA- PCAT7 over-expresses TGF-βR1 expression via binding to miR-324-5p. Moreover, lncRNAs are able to serve as scaffolds. lncRNA-NORAD interacts with importin β1 and increases the interaction of importin β1-SMAD3, contributing to enhanced Smad2/Smad3 expression and nuclear translocation of the SMAD complex phosphorylation, which results in enhancing a number of EMT-related gene expressions. lncRNAs were also found to serve as a guide. lncRNA-ELIT-1, by recruiting SMAD3 to the promoter of TGF-β target genes such as Snail, can act as a positive modulator of TGFβ/SMAD3 signaling and EMT. The canonical Wnt pathway is stimulated when Wnt ligands bind to the Frizzled receptors, which leads to the secretion of β-catenin from the GSK3β–AXIN–APC complex. Then, the secreted β-catenin will be transmitted to the nucleus and binds to TFs TCF or LEF, leading to the activation of EMT-related genes. lncRNAs may serve as signal molecules. lncRNA-AFAP1-AS1 was shown to have the capacity to enhance GSK3β phosphorylation. lncRNA-HOTTIP stimulates β-catenin expression. YY1 transcription factor increases the transcription activity of lncRNA-ARAP1-AS1, which contributes to enhanced EMT via the Wnt/β-catenin signaling pathway. lncRNAs are also able to modulate the canonical Wnt pathway via serving as decoys. lncRNA–H19 and lncRNA-NEAT1 positively regulates the expression of PGRN and CTNB1 via binding to miR-29b-3p and miR-34a-5p, respectively. Moreover, lncRNAs can also act as a guide. The lncRNA–H19 interaction with EZH2 contributes to the Wnt/β-catenin signaling pathway activation, leading to a reduction in the expression of E-cadherin and enhanced tumor metastasis. lncRNA-HOTAIR together with PRC2 has the potential to prohibit WIF-1 expression via stimulating H3K27 trimethylation in its promoter area, whereas they activate the Wnt/β-catenin signaling pathway. The canonical Notch pathway is promoted when the Delta-like or Jagged ligands bind to the Notch receptors. This interaction eventually leads to the secretion of NICD, which exerts its effects on the nucleus. It interacts with some TFs and serves as a transcriptional co-activator to stimulate some EMT–TF expression. lncRNAs were found to function as a guide to mediate the expression of major elements in the Notch signaling pathway. lncRNA-HNF1A-AS1 as well as lncRNA-SNHG12 are capable of over-expressing Notch1 expression. The upregulation of lincRNA-p21 results in the suppression of cancer invasion via downregulating Notch signaling-related proteins, including NICD and Hes-1, and the EMT signaling pathway. Additionally, lncRNAs may serve as a ceRNA to moderate the Notch signaling pathway. lncRNA-UCA1 was shown to be able to enhance JAG1 expression through targeting miR-124. lncRNA-XIST, through targeting miR-137, can enhance Notch1 expression. Growth factors via binding to their receptors concurrently promote the RAS/RAF and PI3K/Akt pathways, leading to the mTOR complex and MEK/ERK signaling axis activation, respectively. The mentioned pathways finally stimulate EMT through inducing some EMT–TF expressions. lncRNAs primarily function as a ceRNA in these pathways. It was shown that lncRNA-UCA1 enhanced CREB1 expression via serving as a ceRNA by targeting miR-582, therefore inducing EMT via the CREB1-mediated PI3K/AKT/mTOR pathway. lncRNA-TTN-AS1 was shown to enhance p-Akt and p-mTOR values likely via targeting miR-497. Additionally, lncRNAs were revealed to serve as signal molecules to regulate Akt and ERK phosphorylation. lncRNA-BANCR enhanced the phosphorylation of MEK and ERK, and lncRNA-ATB is able to enhance Akt and ERK phosphorylation. lncRNA-HOXA-AS3 was shown to be able to increase MEK and ERK phosphorylation via binding miR-29c. This figure was adapted from (223).
3.2.1 lncRNAs and metastasis in ovarian cancer
Wu et al. (2021) examined whether lncRNA GClnc1 was linked to EOC expansion and metastasis (227). They employed RT-qPCR to identify GClnc1 expression in 57 matched EOC and surrounding normal tissue samples. They used GClnc1 silencing and over-expression in SKOV3 and OVC1 cells and measured proliferation, migration, apoptosis, and invasion. They used nuclear or cytoplasmic fractionation protocols, followed by FISH and ISH assays, to determine the subcellular localization of GClnc1. Consequently, they predicted and confirmed the interaction of GClnc1 with forkhead box protein C2 (FOXC2) and FOXC2 with NOTCH1. In EOC tissues, GClnc1 was substantially over-expressed, while GClnc1 knockdown reduced the cells’ viability and increased apoptosis. Furthermore, GClnc1 directly targeted nuclear transcription factor FOXC2 and triggered NOTCH1 transcription. NOTCH1 over-expression increased SKOV3 and OVC1 cell proliferation and EMT and activated the NF-κB/Snail signaling pathway. GClnc1 knockdown also suppressed the metastasis and growth of OVC1 and SKOV3 tumors in the murine model. They concluded that GClnc1 activated the signaling pathway of NF-κB/Snail, boosted the proliferation and metastasis of EOC cell via FOXC2, and increased NOTCH1 transcription (227).
The role of lncRNA cardiac-hypertrophy-associated factor (CHRF) in human cancers and carcinogenesis has been studied—for instance, CHRF was found to be linked with increased colorectal cancer metastasis (228). CHRF was found to regulate the expression of miR-10b, leading to the initiation of EMT, along with increased metastasis and treatment resistance (229, 230). Tan et al. (2020) investigated two ES2 OC cell lines (parental and cisplatin-resistant, CR) and profiled the dysregulated lncRNAs. They found that, most noticeably, CHRF was upregulated in CR ES2 cells. CHRF was considerably increased in OC patients with CR-resistant disease. Patients who had liver metastases were also found to have even higher CHRF levels. Recent research has revealed that miR-10b is involved in madiating cisplatin resistance in OC cells by CHRF. The study found that CHRF increased the resistance to cisplatin in OVCAR, ES2, and SKOV3 OC cells and that this resistance was mediated by EMT and STAT3 signaling activation. EMT and STAT3 activation and cisplatin resistance were all reversed when CHRF was downregulated, but this was abrogated by miR-10b. Then, the findings were confirmed in an in vivo mouse model of cisplatin-resistant EOC, in which miR-10b reduced the effect of CHRF downregulation and lowered the tumor burden. Their findings suggested a new function for lncRNA CHRF in cisplatin-resistant OC. Moreover, CHRF/miR-10b signaling could be a potential therapeutic target (231).
The lncRNA HOTTIP is frequently upregulated in human cancers, where it promotes cancer progression. By sponging miR-216a, lncRNA HOTTIP increased BCL2 expression and chemo-resistance in SCLC (232). HOTTIP increased the expression of PD-L1 in neutrophils, which increased the IL6 levels and promoted the immunological evasion of ovarian carcinoma (233). HOTTIP increased breast cancer cell metastasis, invasion, and EMT (234). Wu et al. (2020) investigated the levels of HOTTIP expression in OC cell lines and clinical tissue samples. The silencing of HOTTIP inhibited ovarian cancer cell rapid growth and invasion as well as migration in vitro, whereas the greater expression of HOTTIP increased invasion in ovarian carcinoma cells, suggesting that HOTTIP could be one of the markers for unsuitable prognosis in OC cases. In addition, HOTTIP acted as a miR-615-3p sponge, thereby increasing the expression of SWI/SNF-associated matrix-linked actin-dependent regulator of the chromatin sub-family E member 1)SMARCE1) (235). Either the upregulation of miR-615-3p or the downregulation of SMARCE1 could abrogate the tumor-promoting effect of HOTTIP in ovarian cancer. Moreover, HOTTIP levels were inversely correlated with miR-615-3p levels and positively correlated with SMARCE1 expression levels in OC cells. HOTTIP knock-out mice showed slower OC xenograft tumor growth in vivo. In conclusion, lncRNA HOTTIP modulates the miR-615-3p/SMARCE1 pathway, thereby enhancing ovarian cancer growth and metastasis (235).
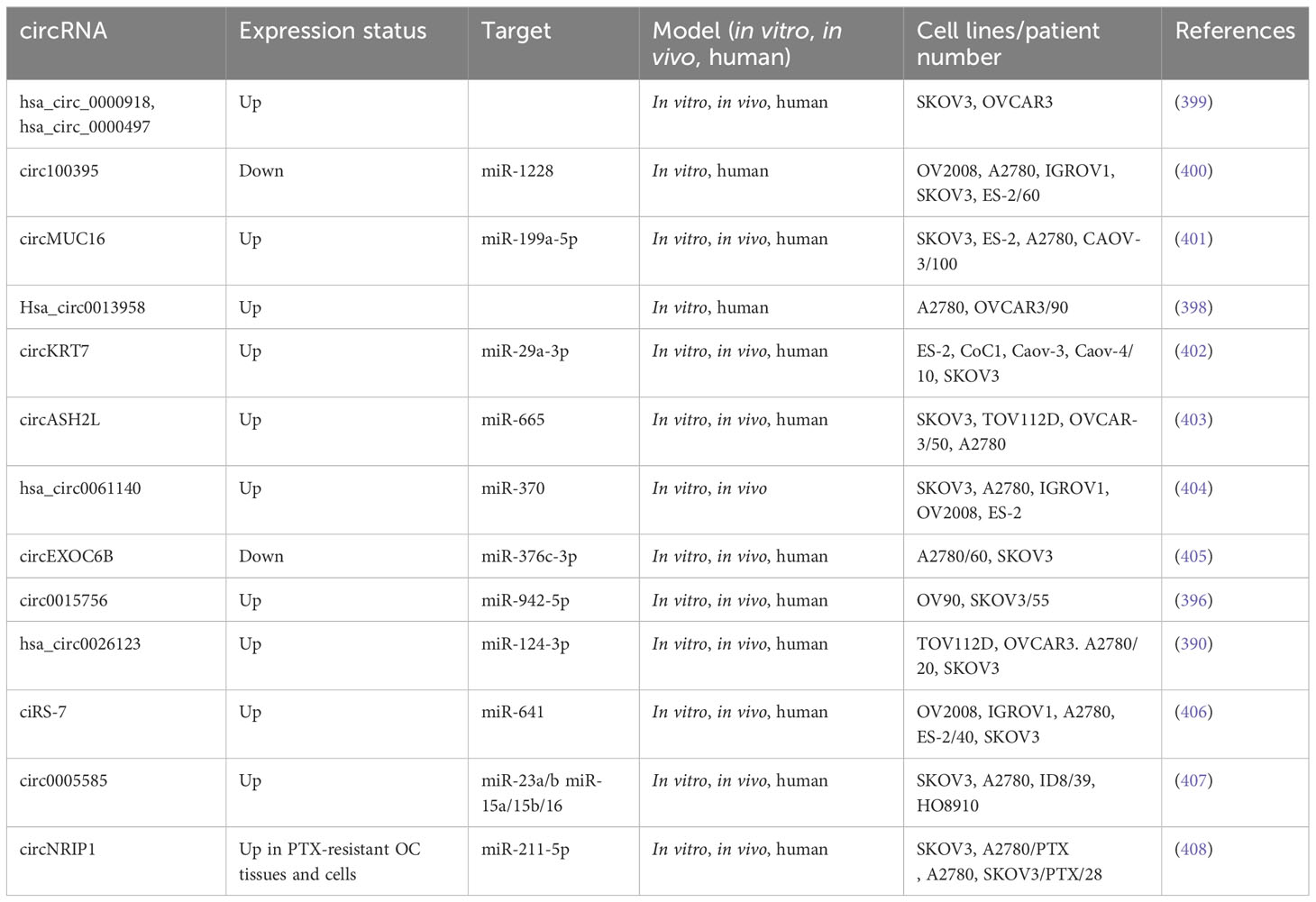
Researchers observed the over-expression of lncRNA EMX2OS in gastric cancer tissues compared to matched control tissue samples (236). AKT3 has been found to promote tumor growth and invasion in seminoma, liver, and thyroid cancer (237). AKT3 was also highly expressed in primary ovarian cancer, and silencing of AKT3 using shRNA considerably reduced the growth of OC cells (238). Duan et al. (2020) explored the expression, cellular function, and mechanism of EMX2OS in OC. RT-qPCR was employed to assess the amounts and activity of EMX2OS in the cell lines and tissues of OC. The relationship between EMX2OS and miR-654 expression in the OC cells was investigated using luciferase and immunoprecipitation assays. Human ovarian cancer tissues were observed to have higher levels of EMX2OS. EMX2OS knock-down decreased OC cell proliferation, spheroid formation, and invasion, whereas the over-expression of EMX2OS showed the opposite effects. Furthermore, EMX2OS promoted tumor development in a human OC xenograft mouse model in vivo. Direct binding of EMX2OS to miR-654 acted as a sponge to downregulate miR-654 and therefore upregulated AKT3, the target of this miRNA. Furthermore, miR-654 reduced cell proliferation, spheroid formation, and invasion, whereas restoration of AKT3 expression counteracted the impact of miR-654 over-expression or EMX2OS silencing. Additionally, in OC cells, PD-L1 was discovered to be a downstream molecule of AKT3 activity. The ectopic expression of PD-L1 in the OC cells abrogated the anti-cancer effects caused by the knock-down of EMX2OS and AKT3 or inducing miR-654 expression. These findings suggest that the EMX2OS/miR-654/AKT3/PD-L1 axis promotes OC malignancy and could be a potential treatment target for this disease (239). Table 4 summarizes some lncRNAs reported to be associated with ovarian cancer metastasis.
3.2.2 lncRNAs and metastasis in endometrial cancer
lncRNA RHPN1-AS1 was found to be over-expressed in several cancer types and is considered to be a cancer promoter (250). Moreover, mitogen-activated protein kinase (MAPK) contributes to the signal transduction from the plasma membranes to the nucleus (285). The ERK pathway is a key type of MAPK involved in numerous processes in cell biology. Importantly, activating the ERK/MAPK pathway may result in EC progression, according to several studies (286). Zhang et al. (2021) explored the role of lncRNA RHPN1-AS1 in the development of EC as well as the associated mechanisms (287). In EC cells and tissues, RHPN1AS1 expression was measured by RT-qPCR, CCK-8, flow cytometry, scratch wound healing, and transwell assays; colony formation has been used as well to measure proliferation, clonogenicity, cell cycle, apoptosis, invasion, and, finally, migration in HEC1A and Ishikawa cells. Moreover, immuno-fluorescence and Western blotting have been used to measure the expression level of protein in Ishikawa and HEC1A cells. They found that RHPN1AS1 expression has been substantially greater in EC cells and tissues. RHPN1AS1 expression in patient samples was linked to the histological grade, FIGO stage, and lymph node metastasis. In Ishikawa and HEC1A cells, silencing of RHPN1AS1 not only inhibited proliferation, cell cycle progression, migration, and invasion but also triggered apoptosis. Furthermore, silencing of RHPN1AS1 decreased Bcl2 expression while increasing the expression of caspase3 and Bax. In addition, MEK and ERK phosphorylation was substantially reduced when RHPN1AS1 was knocked down. The inhibitory effect of silencing RHPN1AS1 on MEK and ERK phosphorylation was further increased after pretreatment with the kinase inhibitor U0126. They concluded that RHPN1AS1 stimulated the ERK/MAPK pathway in EC cells to promote cancer progression while inhibiting apoptosis (287).
The steroid receptor RNA activator (SRA) is a ribonucleoprotein complex-bound functional RNA transcript, which can mediate the co-activation of nuclear steroid receptors. The SRA sequence has a size of ~0.87 kB, with five exons and four introns, and is located on human chromosome 5q31.3. SRA can function as either a ncRNA or protein-coding RNA (288). In the former sense, SRA is a lncRNA that contributes to tumor progression. SRA acts as a molecular coactivator for the genes encoding estrogen and progesterone receptors. SRA has been proven to activate hormone receptors that affect ovarian cancer, breast cancer, and other gynecologic malignancies. lncRNA SRA has been linked to apoptosis, biosynthesis of lipids and steroids, insulin signaling, and muscle development, among several biological processes. Prostate cancer, abnormal cardiac development, and reduced fertility have all been linked to SRA expression (289). Furthermore, one research group investigated the contribution of lncRNA SRA to tumor progression and the associated mechanism. eIF4E-binding protein 1 (eIF4E-BP1) is a downstream mediator of cell proliferation, which could explain the lncRNA SRA mechanism. eIF4E-BP1, one of two major mTOR downstream effectors (290), regulates the expression of several proteins involved in, for example, cell cycle, angiogenesis, cell survival, cancer development, and metastasis at the translational level, thus exerting a critical effect on mTOR signaling. The expression of eIF4E-BP1 is modulated at the transcriptional as well as post-translational levels (291). eIF4E-BP1 is an oncogene which is over-expressed in several cancer types (292). Park et al. (2020) measured SRA expression in EC to establish its biological role and clinical relevance. They tested whether SRA could bind to eIF4E-BP1 and act as a transcription factor by upregulating the Wnt/β-catenin signaling pathway in EC cells and tissues. Consequently, the expression of SRA was higher in EC tissues and cells compared to controls. The transfection of a luciferase reporter plasmid confirmed the binding of SRA to eIF4E-BP1. Furthermore, SRA depletion reduced the expression of eIF4E-BP1 and increased tumorigenesis, EMT, migration, and metastasis. Immunohistochemistry and Western blotting showed that SRA knock-down lowered β-catenin and eIF4E-BP1 expression in the nucleus, whereas SRA over-expression enhanced it. It was concluded that SRA promotes eIF4E-BP1 and Wnt/β-catenin signaling, thus promoting EC proliferation, migration, and invasion. SRA may have a role as one of the prognostic biomarkers as well as a new treatment option in EC (293).
The lncRNA-activated by TGF-β (lnc-ATB) was first found to be upregulated in hepatocellular carcinoma (HCC) (294). lnc-ATB competitively binds to members of the miR-200 family, acting as the regulator of TGF-β signaling, increasing ZEB2 and ZEB1 expression, and promoting EMT as well as invasion in HCC patients. lnc-ATB is now thought to regulate cells’ proliferation or rapid growth, cell cycle, and metastasis and also apoptosis in a variety of other cancers, including osteosarcoma (295). The clinical relevance and mechanism of lnc-ATB in EC were investigated by Zheng et al. (2019). They collected EC samples and normal tissues and identified miRNA targets using bioinformatics analysis (296). In EC cell lines and in a mouse model in vivo, siRNA was used to assess the function of lnc-ATB. lnc-ATB was over-expressed in EC cell lines and tumor tissues. Patients who had a higher level of lnc-ATB expression had a more advanced FIGO stage and poorly differentiated tumors. lnc-ATB interacted with the tumor suppressor miR-126. miR-126 expression was also shown to have a negative correlation with tumor differentiation and FIGO stage. In RL95 and HEC1A cell lines, the knock-down of lnc-ATB resulted in caspase-3-mediated tumor apoptosis as well as G1/S cell cycle arrest by raising the miR-126 levels, leading to decreased cell viability. miR-126 inhibitors affected the expression of the miR-126 target gene PIK3R2 and reversed the cell cycle arrest and tumor inhibition. The knockdown of lnc-ATB increased Sox2-mediated apoptosis. Furthermore, lnc-ATB knock-down reduced the TGFβ-induced EMT phenotype by increasing miR-126 and also decreased migration and invasion.Silencing of lnc-ATB in vivo resulted in a decreased tumor size and a lower expression of PIK3R2/Sox2 and PCNA signaling proteins and reversed the EMT phenotype in the tumor. These findings showed that lnc-ATB suppressed miR-126 and therefore acted as a tumor promoter in EC (296).
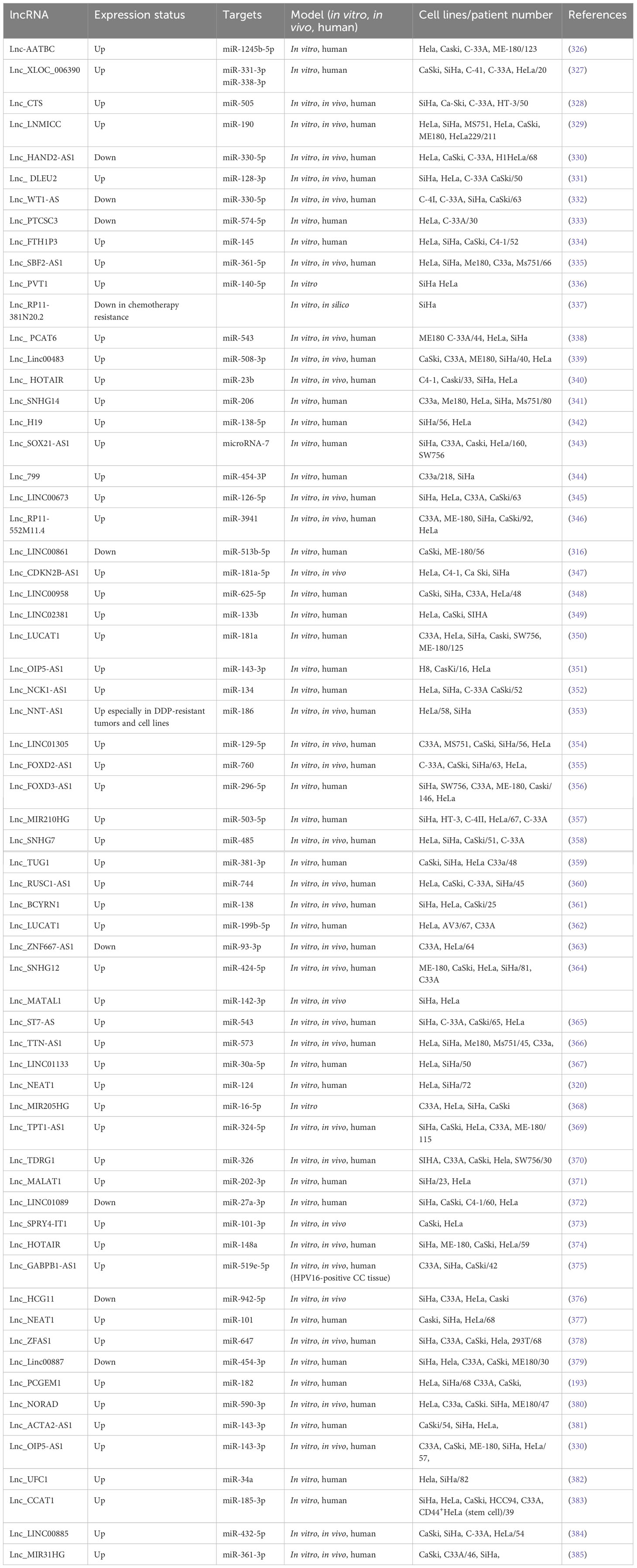
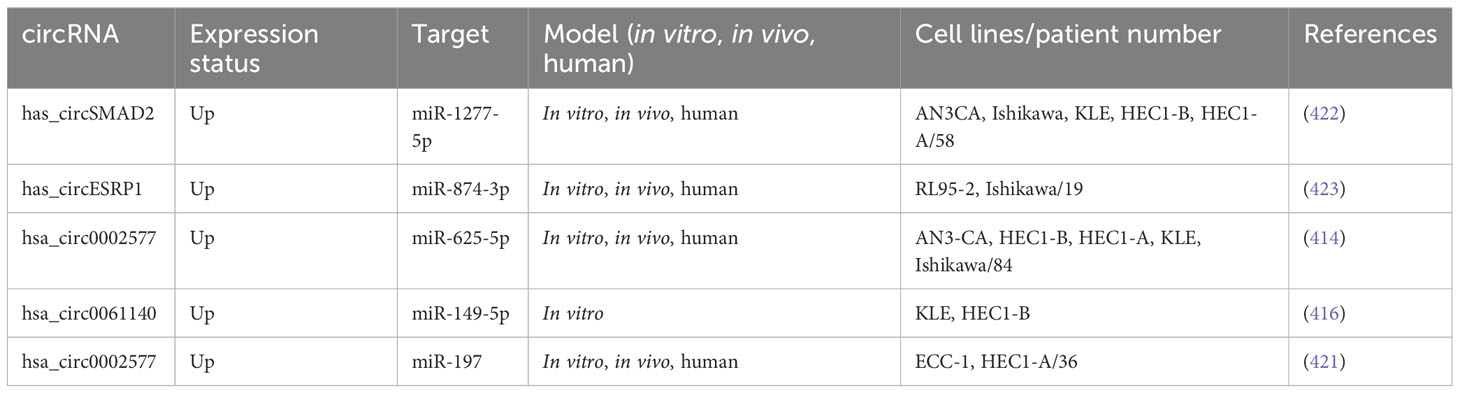
lncRNA HOTAIRM1 was observed to be expressed in myeloid cells, the exact location of which was later found to be on human chromosome 7p15.2 (297). In fact, HOTAIRM1 controls the expansion of the cell cycle during the maturation of myeloid precursor cells and is upregulated in NB4 human promyelocytic leukemia cells as well as in myeloid leukemia patients (298). HOTAIRM1 is also involved in the progression of several other cancers, such as breast cancer, pancreatic ductal adenocarcinoma, and glioma (299). Anti-sense lncRNAs are transcribed from the opposite strand of genes, encoding proteins or are non-protein coding, and are strongly linked to tumor progression (300). Moreover, HOTAIRM1 is situated at the 5′ end of homeobox A (HOXA) gene cluster in an anti-sense manner and contains a similar CpG island as the HOXA1 starting point (297). HOTAIRM1 has been shown to increase HOXA1 expression in myeloid-derived lung cancer suppressor cells and in glioblastoma multiforme (301). HOXA1 is a member of the HOX gene family, which is composed of four gene clusters (HOXA, HOXB, HOXC, and HOXD) that play important roles in regulating embryonic development and cell differentiation. HOXA1 is highly expressed in several types of cancer, including breast cancer, oral squamous cell carcinoma, hepatocellular carcinoma, and gastric cancer, and is associated with a poor prognosis. Studies have shown that HOXA1 plays a key role in regulating the cell cycle, promoting EMT, and enhancing tumor cell proliferation, migration, and invasion. As such, HOXA1 is considered to be a cancer-promoting gene (302). Li et al. (2019) explored whether HOTAIRM1 and the respective sense transcript HOXA were involved in carcinogenesis and expansion of type I EC. They applied Western blotting and qRT-PCR to determine HOXA1 and HOTAIRM1 expression levels in the type I EC tissues. Additionally, in vitro and in vivo, gain-and-loss-of-function studies have been performed to examine the biological roles of HOXA1 and HOTAIRM1 in type I EC. Type I EC tissues were found to have considerably higher levels of HOTAIRM1 and HOXA1. Moreover, HOTAIRM1 and HOXA1 expression was shown to be linked to lymph node metastasis, FIGO stage, and also with each other. Proliferation, migration, invasion, and EMT were dramatically reduced when HOTAIRM1 was knocked down, and the opposite effects were seen when HOTAIRM1 was upregulated. Furthermore, they discovered that HOTAIRM1 affected HOXA1 gene expression in type I EC cells. Furthermore, HOXA1 knockdown inhibited cancer progression, thereby confirming HOXA1 to be an oncogene. Moreover, the involvement of HOXA1 and HOTAIRM1 in promoting tumor development in vivo was validated. They showed for the first time that HOTAIRM1 regulated HOXA1 in the type I EC by acting as the oncogene. The HOTAIRM1/HOXA1 axis may not only be a predictive biomarker but also a therapeutic target in type I EC (303). Table 5 shows a list of some lncRNAs, which have been reported to be linked to metastasis in endometrial cancer.
3.2.3 lncRNAs and metastasis in cervical cancer
Recent studies have suggested that the intergenic long non-coding RNA (lncRNA) LINC00861 may play a role in improving the prognosis of several types of cancer. In particular, the downregulation of LINC00861 has been linked to poor outcomes in ovarian cancer patients (268). In CC, researchers observed that lncRNAs, such as colon cancer-related transcript-1 and plasmacytoma variant, act as ceRNAs in order to remove miRNAs that promote EMT (315). Liu et al. (2021) designed a study for investigating the involvement and underlying mechanisms of LINC00861 in the development of ovarian cancer (316). RT-qPCR was employed for measuring LINC00861 and miR-513b-5p expression. CCK-8, transwell, and colony formation assays were utilized for measuring viability and proliferation as well as migration. To verify whether miR-513b-5p targeted LINC00861 and PTEN, the researchers utilized a luciferase assay, while Western blotting was applied to measure the expression of proteins. They demonstrated LINC00861 expression in the CC tissues. ME180 and CaSki cell lines were considerably lower compared to controls. The downregulated LINC00861 expression levels were linked to an advanced stage, poor survival, and lymph node metastasis in CC patients. The PI3K/Akt/mTOR signaling pathway was substantially enhanced in CC samples with low LINC00861 expression levels, compared to CC samples with high LINC00861 expression levels, according to Gene Set Enrichment Analysis. The over-expression of LINC00861 suppressed the CC cells’ proliferation, migration, invasion, and EMT and the phosphorylation of Akt and mTOR proteins, while it increased PTEN protein expression. A dual-luciferase reporter gene assay has been employed to confirm the interconnection of LINC00861, PTEN, and miR-513b 5p. In both cell lines, the level of PTEN expression has been remarkably lower in the cells given treatment with a miR-513b 5p mimic, while this has been substantially greater in the cells treated with a miR-513b 5p inhibitor in comparison to a control NC mimic and a control NC inhibitor. Moreover, LINC00861 was found to sponge miR-513b-5p and further enhance PTEN expression in CC cells, suggesting its possible function as a competitive endogenous RNA. The cells that have been co-transfected with the miR-513b 5p and LINC00861 mimics showed a significant increase in PTEN expression, Akt and mTOR phosphorylation, and the EMT phenotype. The LINC00861/miR-513b 5p axis could inhibit the progression of CC and limit the EMT process by regulating the PTEN/Akt/mTOR signaling pathway (316).
The lncRNA nuclear-rich transcript 1 (lncRNA-NEAT1) stimulates the proliferation and invasion of CC cells while inhibiting apoptosis (317). One study investigated the putative mechanisms of lncRNA-NEAT1 in CC. Prior investigations have found a major contribution of miR-124 to various types of cancer (318). Therefore it was hypothesized that lncRNAs could influence tumor growth by functioning as a molecular sponge for miR-124, thus regulating the expression of target mRNAs (319). The contribution of lncRNA-NEAT1 and its sponging of miR-124 to CC progression, as well as the associated mechanisms, was examined by Shen et al. (2020). They investigated the relationship between lncRNA-NEAT1 expression with CC patient clinical features. In addition, researchers measured migration and invasion using transwell and scratch wound healing assays. In addition, anchorage-independent colony formation assays and CCK-8 have been used to measure cell growth. TargetScan, RNA pull-down assays, and, finally, dual-luciferase reporter gene served to predict and validate the binding of miR-124 to lncRNA-NEAT1. Moreover, researchers applied Western blotting to measure MMP-2, MMP-9, and NF-κB pathway-associated factors and EMT-related factors (vimentin, E-cadherin, and N-cadherin). The lncRNA-NEAT1 expression elevated in the CC tissues and cells with a positive correlation to lymph node metastasis and TNM stage in the patients. When lncRNA-NEAT1 was over-expressed in SiHa or HeLa cells, proliferation, migration, invasion, and the NF-κB pathway were enhanced, and the EMT markers were altered. The opposite effects were observed when lncRNA-NEAT1 was knocked out. Furthermore, the impact of lncRNA NEAT1 on HeLa cell motility, EMT, invasion, and the NF-κB pathway was abrogated by the administration of miR-124. They concluded that lncRNA-NEAT1 modulated the miR-124/NF-κB pathway, thereby promoting CC cell invasion and dissemination (320).
NF-κB-interacting lncRNA (NKILA) is located on chromosome 20q13 and modulates the signaling pathway involving inhibitory protein IκB kinase (IKK) and NF-κB. The NKILA expression levels were illustrated to be inversely correlated to the invasion of breast cancer and metastasis. NKILA has been observed to be downregulated in ESCC tissues and cancer cells. In addition, NKILA inhibited the signaling of NF-κB to hinder ESCC cells’ migration and rapid growth. The inhibitory protein IKK keeps NF-κB in an inactivated state in the cytoplasm by forming a trimer and prevents the nuclear translocation of the NF-κB transcription factor (321). Furthermore, NF-κB was discovered to be regulated in a negative feedback loop because it increases NKILA expression, thereby creating a NF-κB/NKILA complex to suppress NF-κB activation in normal mammary epithelial cells (322). As a result of the reciprocal feedback loop of NKILA and NF-κB, lncRNAs may bind to various components of the pathway in order to regulate signaling.
Chronic inflammation contributes to the metastasis and invasion of CC, and NF-κB signaling is known as a key connection of inflammation with tumor growth (323). Wang et al. (2020) addressed the impact of NKILA on metastasis and proliferation and the associated mechanisms in CC cell lines (324). The NKILA expression levels were determined in vitro and in vivo using RT-qPCR. CaSki cells were transfected with a short hairpin RNA targeting NKILA and an appropriate control, whereas C33A cells were transfected with an over-expression vector, pcDNA3.1NKILA, and a control sequence. CCK-8, Western blotting, Matrigel invasion, and scratch wound healing assays were used to evaluate migration, proliferation and invasion as well as EMT expression in C33A and CaSki cells. NKILA expression is lower in the CC cell lines (C33A, SiHa, HeLa, and CaSki) and tissue samples. The downregulation of NKILA expression using shRNA dramatically increased CC cells’ proliferation, which increased the invasion in C33A cells. The upregulation of NKILA reduced the invasion, migration, and proliferation of the CaSki cells. As shown by measurements of E-cadherin, vimentin, ZO-1, and N-cadherin, it has been suggested that NKILA could inhibit the EMT to lessen the potential for metastasis. In addition, the knockdown of NKILA enhanced the breakdown of IKK and promoted the nuclear translocation of p65 in tC33A cells. By contrast, NKILA over-expression reduced NF-κB activation in CaSki cells. They concluded that NKILA was linked to NF-κB activation and could modulate EMT processes to reduce invasion and migration in CC cells (324).
Recent studies have suggested that intergenic lncRNA 518 (LINC00518), located on chromosome 6, dysregulated in melanoma and triple-negative breast cancer. Wang et al. (2019) analyzed the expression pattern, biological function, and clinical relevance of LINC00518 in CC (325). Moreover, flow cytometry has been employed for detecting cell apoptosis, and MTT and colony formation assays have been applied for measuring proliferation or rapid growth, whereas scratch wound healing and transwell assays were employed to assess invasion and migration. In addition, the expression of EMT markers and JAK/STAT3 signaling proteins was detected using Western blotting. LINC00518 was found to be over-expressed in CC tissues with an association with lymph node metastasis, FIGO stage, cervical invasion depth, and poor prognosis in CC cases. LINC00518 has been shown to be a potent, independent prognostic marker for the overall rates of survival, according to univariate and multivariate Cox regression analyses. The analysis demonstrated the inhibition of migration and proliferation as well as invasion and increased apoptosis following LINC00518 silencing in vitro. LINC00518 silencing also suppressed the N-cadherin and vimentin levels via inhibiting JAK/STAT3 activation. LINC00518 was found to operate as the oncogene in CC via the regulation of the JAK/STAT3 signaling pathway and may have a role as a prognostic biomarker and a possible therapeutic target (325). Table 6 shows a list of some metastasis-related lncRNAs in cervical cancer.
3.3 circRNAs and metastasis in gynecological cancer
3.3.1 circRNAs and metastasis in ovarian cancer
The circRNA vacuolar protein sorting 13 homolog C (circVPS13C) has been found to be upregulated in ovarian cancer (386). However, the cellular mechanisms by which circVPS13C promotes ovarian cancer were unclear. In one study, miR-145 influenced Sp1 and Cdk6 levels to increase paclitaxel sensitivity in ovarian cancer cells (387). Nevertheless, the mechanism by which propofol could mediate miR-145 suppression of ovarian cancer cells was still unclear. Lu et al. (2021) reported that cell cycle, survival, and metastasis of ovarian cancer cells were inhibited, while apoptosis was increased, after propofol administration (388). It was discovered that propofol affected CircVPS13C and miR-145 to act against OC. MTT and transwell assays have been used to measure the survival and metastasis of ovarian cancer cells. Flow cytometry has been employed for studying apoptosis and the cell cycle. In addition, miR-145 and circVPS13C expression levels were measured using RT-qPCR. Moreover, the circinteractome database predicted a target binding between miR-145 and circVPS13C, which was later confirmed using RNA pull-down assay and dual-luciferase reporter assay as well as RNA-binding protein immuno-precipitation (RIP). In addition, the levels of ERK, p-ERK, MEK, and p-MEK in the OC cells were determined using Western blotting. Treatment with propofol reduced the survival, migration, and cell cycle of the OC cells while increasing apoptosis. The miR-145 levels were dose-dependently increased by propofol, which explained its anti-cancer activity. circVPS13C also directly targeted miR-145. Propofol inhibited ovarian cancer development by decreasing circVPS13C, leading to an increase in miR-145. In conclusion, propofol affected the circVPS13C/miR-145/MEK/ERK signaling pathways for inhibiting malignant properties and upregulating apoptosis in ovarian cancer cells (388).
Several types of cancers (e.g., hepatocellular carcinoma, bladder cancer, and EC) can be effectively inhibited by miR-124-3p (389). Yang et al. (2021) explored the role of hsa-circ0026123 in vitro and in vivo. They used a luciferase reporter assay to investigate the relationships between miR-124-3p, EZH2, and hsa-circ0026123. They analyzed protein and gene expression with Western blotting and RT-qPCR. Nude mouse tumor xenografts generated from SKOV3 cells were used to evaluate tumor growth after regulation of hsa-circ0026123. OC tissues and cell lines displayed higher expression levels of hsa-circ0026123 compared to controls, whereas silencing of hsa-circ0026123 suppressed proliferation, migration, and differentiation markers in cancer stem cells (CSC). Rescue studies as well as the luciferase reporter assay demonstrated that the downregulation of hsa-circ0026123 led to the sponging of miR-124 3p and further suppression of EZH2. They concluded that hsa-circ0026123 affected the miR-124-3p/EZH2 signaling pathway to suppress ovarian cancer, and this approach may be one of the potent bio-markers for OC and possibly a target proposed for treatment (390).
Researchers have shown that hsa-circ0015756 was substantially over-expressed in OC tissues (391). miR-942 in OC tissues was noticeably lower compared to healthy controls (392), and its over-expression accelerated the aggressiveness of melanoma by inhibiting DKK3 (393). CUL4B is a constituent of Cullin4B-Ring E3 ligase scaffold protein complex (394). CUL4B works as an oncogene in diverse kinds of cancer and is also over-expressed in OC tissues, leading to alterations in CDK2 and cyclin D1 levels and further increases in proliferation (395). Du et al. (2020) designed an experiment to analyze the involvement of circ-0015756 in OC and the associated pathways. Moreover, they used Western blotting as well as RT-qPCR to measure miR-942-5p and CUL4B as well as circ-0015756. Flow cytometry, colony formation, CCK-8, and transwell assays have been used to measure apoptosis, invasion, proliferation, and migration. In fact, Western blotting test has been applied to measure the amount of proteins involved in proliferation and metastasis. RNA pull-down assay and RNA immunoprecipitation assay as well as dual-luciferase reporter assay have been used to demonstrate the interactions of miR-942-5p, circ-0015756, and CUL4B. Tumor development in vivo was measured in a mouse xenograft model. The levels of CUL4B and circ0015756 were higher and the miR-942-5p levels were lower in OC cells and tissues compared to controls. The depletion of circ-0015756 in OC cells suppressed the migration, invasion, and proliferation during apoptosis development. The depletion of circ-0015756 increased miR-942-5p, thereby inhibiting OC cell growth. The upregulation of miR-942-5p lowered CUL4B and inhibited OC cell growth. They concluded that circ-0015756 sponged miR-942-5p to increase the expression of CUL4B and promote OC progression. Furthermore, the suppression of circ-0015756 reduced tumor progression in vivo and could be a possible treatment for OC (396).
hsa-circ0013958 was shown to affect the development of NSCLC via miRNA134 sponging, leading to the over-expression of cyclin D1 (397). Nevertheless, the role of hsa-circ0013958 in ovarian cancer and the possible mechanisms needed further clarification. hsa-circ0013958 was upregulated in OC cells and tissues and acted as an oncogene, according to a study by Pei et al. (2020). In their study, RT-qPCR has been employed to measure the hsa-circ0013958 level in 45 pairs of matched OC cells and tissues, and the clinicopathological relevance and diagnostic value were determined. CCK-8 test and transwell assay as well as flow cytometry have been employed to measure the migration, proliferation, invasion, and apoptosis of OVCAR3 and A2780 cells. Western blotting was used to measure the apoptosis-associated proteins Bcl2 and Bax and the EMT-associated proteins E-cadherin and vimentin. hsa-circ0013958 was found to have an abundant expression in OC tissues and cells, with an association to the patient’s lymph node metastasis and FIGO stage. The in vitro knock-down of hsa-circ0013958 suppressed OC proliferation or rapid growth, migration, and invasion and increased apoptosis. Both EMT and apoptosis-associated proteins were significantly altered. To conclude, hsa-circ0013958 may influence EMT and apoptosis and contribute to OC progression (398). Table 7 shows a list of contributions of some metastasis-related circular RNAs to ovarian cancer.
3.3.2 circRNAs and metastasis in endometrial cancer
The blood levels of hsa-circ0002577 in EC patients were found to be 2.4 folds greater than in the healthy females, whereas the other circRNAs that were examined varied from 1.43 to 2.05 folds higher in healthy women (409). The WDR26 gene is a precursor of hsa-circ0002577. WDR26 was over-expressed in malignant breast tumors, resulting in PI3K/Akt pathway activation and further progression and spread of breast cancer (410). Accordingly, the hsa-circ0002577 upregulation in EC might inhibit tumor formation. A variety of intracellular signaling pathways, including MAPK signaling, can recruit IGF1R (a transmembrane tyrosine kinase receptor), and PI3K/Akt is an important participant in this pathway (411). IGF1R over-expression was found to be linked to a worse prognosis in EC cases, and the IGF1R expression level was significantly higher in the developed EC tissues in comparison with the early stage or the proliferative endometrial samples (412). IGF1R monoclonal antibodies and IGF1R-selective inhibitors are being tested for their abilities to suppress tumor metastasis and progression while also increasing tumor susceptibility to other biological treatments (413). Wang et al. (2020) explored whether hsa-circ0002577 regulated EC progression (414). They collected tumor samples and surrounding normal tissues from 84 EC patients. The EC cells have been transfected with miR-625-5p mimics, lentiviral vectors that expressed IGF1R, a miR-625-5p inhibitor, recombinant lentiviral vectors expressing hsa-circ0002577 (Lv-circRNA), short hairpin RNAs against hsa-circ0002577 (sh-circRNA), and their specific controls. Ishikawa cells that had been transfected with the sh-circRNA or a control sequence were inoculated into a BALB/c mouse to produce a xenograft model. In comparison to normal controls, the researchers observed the expression of hsa-circ0002577 in EC cells as well as tissue samples. They also showed that there was a relationship between hsa-circ0002577 expression and poor prognosis and more advanced stage in EC patients. Lv-circRNA-transfected EC cells showed increased proliferation, migration, and invasion, while sh-circRNA-transfected cells showed the opposite effects. In EC cells, hsa-circ0002577 functioned as a miR-625-5p sponge. Moreover, IGF1R has been identified as one of the possible downstream targets of miR-625-5p. IGF1R expression was higher in the EC tissues compared to controls and was shown to stimulate the PI3K/Akt signaling pathway. hsa-circ0002577 increased IGF1R expression and the PI3K/Akt signaling pathway activity. Mice inoculated with hsa-circ0002577 knockdown tumor cells showed slower tumor development and less metastasis. They proposed that hsa-circ002577 could be a promising therapeutic target to treat EC (414).
According to the studies, hsa-circ0061140 promotes OC expansion and spreads via sponging miR-370 (404). miR-149-5p increased the expression of ARF GTPase-activating protein (GIT1) in order to inhibit the development of medullary thyroid cancer cells (415). The study of Liu et al. (2020) addressed the impacts of hsa-circ0061140 on EC progression. hsa-circ0061140 knockdown slowed the proliferation of EC cells by affecting the miR-149-5p and STAT3 axis. Functional assays demonstrated that the downregulation of hsa-circ0061140 abrogated its sponging activity for miR-149-5p and suppressed the EC cells’ development. STAT3 has been revealed as the miR-149-5p downstream target gene. In addition, miR-149-5p has been widely linked to tumor development and dissemination. The direct binding of hsa-circ0061140 to miR-149-5p has been shown by RIP assays and a dual-luciferase reporter. The expression of STAT3 has been shown to be downregulated by miR-149-5p. They discovered that hsa-circ0061140 exerts its oncogenic effect by regulation of the STAT3 and miR-149-5p axis and might play a role in EC therapy (416).
hsa-circ0002577 was found to be upregulated in specimens of EC patients (409). In contrast, it was found to be downregulated in CC. When upregulated, it targeted FOXM1, resulting in the suppression of proliferation and invasion (417). Catenin delta 1 (CTNND1) is also called p120-catenin, which has been first discovered as a substrate of the oncogenic tyrosine kinase Src (418) and later found to be a constituent of the adherens junction complex containing E-cadherin and catenin proteins (α, β, and γ) (419). CTNND1 may be promising for presenting novel therapeutic options in the future—for example, miR-298 suppressed HCC progression via blocking CTNND1-mediated Wnt/-catenin signaling (420).
Shen et al. (2019) studied the possible role of hsa-circ0002577 in EC development. They showed that hsa-circ0002577 expression is considerably higher in EC tissues, which was associated to the FIGO stage, lymphovascular invasion, and a worse overall prognosis in EC patients. The EC cells’ proliferation, invasion, and migration in vitro as well as tumor development in vivo have all been suppressed when hsa-circ0002577 was silenced. Mechanistic investigations suggested that hsa-circ0002577 may function as the sponge for miR-197. Moreover, CTNND1 has been found as a miR-197 target gene. They also discovered the oncogenic impacts of hsa-circ0002577 mediated by regulating the miR-197/CTNND1/Wnt/β-catenin axis (421). Table 8 reports several metastasis-related circular RNAs involved in endometrial cancer.
3.3.3 circRNAs and metastasis in cervical cancer
miR-1270 enhanced the proliferation and metastasis of osteosarcoma cells, and over-expression of miR-1270 was linked to poor survival in osteosarcoma patients (424). CircCdr1 has been shown to inhibit miR-1270 expression and promote SCAI expression, thereby enhancing cisplatin sensitivity in ovarian cancer (425). The transcription factor ZEB2 (426) has several roles in both pathological and physiological processes, such as neurological development and preservation of macrophage tissue specificity, and also in carcinogenesis (427). ZEB2 upregulates MMP activity and reduces E-cadherin epithelial marker and intercellular adhesion, thus facilitating tumor cell invasion (428). ZEB2 was found to be abundantly expressed in CC cells, where it promoted EMT and metastasis (429). Wang et al. (2021) found significantly higher expressions of circ0001247 in the CC cells and tissues. circ0001247 could regulate the miR-1270/ZEB2 axis to promote CC cell proliferation and dissemination as well as invasion while also inhibiting apoptosis. In addition, circRNA expression in the CC and normal cervical cell lines was obtained from GEO database (GSE147483 dataset), and circ0001247 was found to be the most distinct circRNA. RT-qPCR has been employed to measure miR-1270 and ZEB2 expression in vitro and in vivo. In addition, the binding of circ0001247 to miR-1270, as well as the binding of miR-1270 to 3′UTR of ZEB2, was confirmed using dual-luciferase reporter gene assays. GSE147483 analysis showed that circ0001247 could function as an oncogenic circRNA in CC. circ0001247 expression in the CC cell lines and tissues has been greater in comparison to the healthy cervical epithelial cells and surrounding normal tissue. Silencing of circ0001247, as well as over-expression of miR-1270, promoted proliferation and metastasis while inhibiting apoptosis in CC cells. Furthermore, circ0001247 was found to sponge miR-1270 and increase ZEB2 expression to accelerate CC development (430).
Multiple myeloma and intrahepatic cholangiocarcinoma were shown to have lower levels of circSMARCA5 (431, 432), whereas bladder and breast cancer had higher levels (433). circSMARCA5, therefore, seems to perform a variety of functions in different cancers. The expression of circSMARCA5 was shown to be lower in CC (434). Tudor Staphylococcal Nuclease or p100 protein (SND1) was first identified as an Epstein–Barr virus nuclear protein 2 co-activator and is an example of a staphylococcal nuclease domain-containing protein. The SND1 protein regulates pre-mRNA splicing as well as gene transcription and contributes to the formation and progression of different cancers. SND1 protein has also been linked to cervical cancer metastasis (435). The 14-3-3 subtype of the YWHAB protein is involved in cell redox metabolism, apoptosis, cell cycle, and autophagy along with several other physiological processes (436). Zhang et al. (2021) analyzed the role of circSMARCA5 in CC development. They used RT-qPCR to show that the expression of SMARCA5 was lower in CC cells and tissues. The over-expression of SMARCA5 in CC cells reduced proliferation and invasion while promoting apoptosis, as shown by transwell, Annexin V-FITC PI detection kit, and CCK-8 assays. Western blotting was used to measure apoptosis-associated proteins. Moreover, interaction of SND1 with SMARCA5 has been suggested by StarBase and confirmed by an RNA pull-down experiment. STRING was used to predict the protein interactions of SND1 and SMARCA5, which was confirmed by a co-immunoprecipitation experiment. In addition, loss-and-gain-of-function investigations have been employed to determine the effects of SND1 or YWHAB on CC progression. Knockdown of SND1 or YWHAB was found to offset the effects of short interfering RNA to target SMARCA5 on the migration, apoptosis, invasion, and rapid growth of CC cells. SMARCA5 upregulation inhibited CC metastasis in vivo. circSMARCA5 upregulation increased apoptosis in CC cells, while it suppressed SND1 binding to YWHAB and reduced proliferation, invasion, and metastasis in CC (437).
It has been suggested that circUBAP2 might be a prognostic indicator due to its contribution to various malignancies, such as osteosarcoma, triple-negative breast cancer, and lung cancer (435). It was recently shown that miR-361-3p levels declined in CC patient samples. Moreover, greater levels of miR-361-3p were an independent predictor of better outcomes (438). SOX4, a SOX transcription factor family member, was upregulated in CC, leading to progression and treatment resistance (439). Several investigations have reported the possible role of miR-361-3p and SOX4 in CC carcinogenesis. Meng et al. (2020) examined the expression pattern of circUBAP2 and the underlying mechanisms of action (440). They measured the level of circUBAP2, N-cadherin, miR-361-3p, vimentin, SOX4, Bax, cleaved caspase 3, Bcl-2, and E-cadherin using RT-qPCR and Western blotting. MTT assay and flow cytometry as well as transwell assay have been employed to measure the apoptosis, rapid growth or proliferation, invasion, and migration of CC cells. A luciferase reporter assay and a pull-down test demonstrated the relationship of miR-361-3p with circUBAP2 or SOX4. A murine xenograft model has been created by injection of SiHa cells that were stably transfected with sh-circUBAP2. In addition, circUBAP2 has been found to be over-expressed in CC cells and tissues, and high levels of circUBAP2 predicted poor outcomes in patients. circUBAP2 knockdown triggered apoptosis in vitro and suppressed proliferation, invasion, migration, and EMT. The knockdown of circUBAP2 inhibited metastasis and tumor growth in vivo. Moreover, miR-361-3p could directly bind to both circUBAP2 as well as SOX4 mRNA, suggesting that circUBAP2 is capable of regulating the expression of SOX4 via miR-361-3p sponging in CC cells. Moreover, rescue experiments showed that miR-361-3p downregulation or SOX4 over-expression in CC partly reversed the circUBAP2 knockdown-induced stimulation of cell growth and metastasis. Since circUBAP2 promotes CC tumor metastasis and expansion via affecting the miR-361-3p/SOX4 axis, it may be a potent CC treatment target and prognostic marker (440).
The targeting of APC regulators of the Wnt signaling pathway by miR-218 was discovered to inhibit CC cell progression (441). miR-218 has been shown to inhibit several cancers such as ovarian, bladder, and prostate (442). HOXA1 is considered to be an oncogene that promotes proliferation, invasion, and metastasis. The upregulation of HOXA1 has been linked to worse survival rates in CC patients (443). Mao et al. (2019) discovered that CC cell lines and tissues had substantially higher levels of circEIF4G2. In addition, higher circEIF4G2 levels were linked to a worse outcome in CC patients. Moreover, rapid growth of cells, colony formation, and invasion as well as migration were all reduced when circEIF4G2 was knocked down in CC cells. circEIF4G2 was also discovered to act as a sponge for miR 218, which, in turn, was known to target HOXA1 mRNA. Therefore, circEIF4G2 could sponge miR-218 to increase the expression levels of HOXA1. Transfection with a miR-218 inhibitor abrogated the inhibitory impact of circEIF4G2 knockdown on cell invasion, proliferation, and migration, according to rescue studies. Moreover, the impact of the miR-218 inhibitor on CC cells was also reversed when HOXA1 was silenced. Hence, circEIF4G2 boosted cell proliferation and migration through the miR-218/HOXA1 pathway (444).
miR-320a has been shown to increase proliferation, invasion, migration, and chemosensitivity and inhibit apoptosis in various cancer cells, such as salivary adenoid cystic carcinoma, liver cancer, and some other cancers (445). Nevertheless, miR-320a’s contribution to CC was only demonstrated by one study (446). In a number of human malignancies, FOXM1 was shown to increase proliferation, invasion, migration, and EMT (447). A correlation has been observed between FOXM1 and Bcl-2 and Ki-67 expression, as well as enhanced gastric cancer cell proliferation (448). FOXM1 increased E-cadherin, caveolin-1, uPA receptor (uPAR), and urokinase-type plasminogen activator (uPA) to induce cell EMT (449). miR-320a was found to directly target FOXM1 and therefore could inhibit survival, migration, and invasion (450). Some reports about metastasis-related circRNAs involved in cervical cancer are listed in Table 9.
4 Conclusions
This review highlights the important role of non-coding RNAs, including microRNAs, long non-coding RNAs, and circular RNAs, in the metastasis of gynecological cancers. ncRNAs have been demonstrated to contribute to all stages of metastasis in most types of cancers, controlling proliferation, migration, invasion, EMT, and metastasis. These molecules regulate various aspects of the metastatic process, including cellular transformation, tumor growth, invasion, migration, and angiogenesis. Additionally, they can act as prognostic markers and potential therapeutic targets for gynecological cancers. There are complex interactions between ncRNAs and proteins, DNA, and complementary RNA molecules to affect metastasis, as might be expected given the complexity of the metastatic process. To further understand the role of ncRNAs and the affected signaling networks in metastasis, powerful gene function-based methods are required. Rapid sequencing of the human genome (including ncRNAs) is now possible through the latest advancements in genome editing techniques like CRISPR/Cas9 technology. Combining functional genetic screening with appropriate animal models and single-cell-based assays is now within reach, which will enable us to better understand the molecular processes controlling the function of ncRNAs in metastasis. Moving forward, there are several avenues for future investigation. First, further studies are needed to elucidate the molecular mechanisms by which non-coding RNAs contribute to the metastatic process. This will provide a better understanding of how these molecules can be targeted for therapeutic purposes. Second, the development of non-invasive diagnostic methods for gynecological cancers based on non-coding RNAs is an important area for future research. Third, the identification of novel non-coding RNAs that play a role in gynecological cancer metastasis will provide new targets for therapeutic intervention. Fourth, the use of non-coding RNAs as therapeutic agents in the treatment of gynecological cancers is an exciting prospect that warrants further investigation. Moreover, the roles of ncRNAs in gynecologic cancer progression will require further validation by analyzing sufficient numbers of clinical samples. ncRNAs are likely to become biomarkers for the diagnosis and prognosis of gynecologic cancers when their specific expression levels have been sufficiently validated in these cancers. Furthermore, the development of new drug delivery methods will be necessary to employ ncRNAs as therapeutic targets and anticancer agents.
Noteably, there is no single non-coding RNA (ncRNA) that plays a major role in gynecological cancer metastasis. Rather, several ncRNAs, including microRNAs, long non-coding RNAs, and circular RNAs, have been shown to play important roles in regulating various aspects of the metastatic process in gynecological cancers. The specific ncRNAs involved can also vary depending on the type and subtype of gynecological cancer.
It seems that a combination of several ncRNAs, rather than a single one, is involved in the metastasis of cancers. Further research in this area is needed to fully understand the specific roles of different ncRNAs in gynecological cancer metastasis and to identify potential therapeutic targets.
Table 10 contains a summary of miRNA and lncRNA data in metastatic gynecological cancers. Due to conflicting reports regarding the function of miRNA in different cancers (upregulation or downregulation), we have combined the data of the three cancers studied in different studies (at least two studies) to determine the percentage increase or decrease in expression. Accordingly, we have divided miRNA’s possible roles into three general categories: miRNAs that were reduced in all studies (100%) as miRNAs with tumor suppressor potential and, in contrast, miRNAs with increased expression in all studies as miRNAs that have oncomiR potential. The third category is miRNAs, which are located between these two categories and are in the unknown category. Further studies are needed to determine their role. In Table 10, in addition to the up and down percentages, we also provide the number of studies on which this percentage has been calculated. As a result, the greater the number of studies, the more reliable the role of miRNA (tumor suppressor or oncomiR) is, based on up and down percentages—for example, miR-218 is a potent tumor suppressor with the highest number of reports of downregulation in various studies and simultaneous targeting of 10 critical genes in cancer, so, further studies to evaluate the therapeutic application of this miRNA in gynecological cancers could be valuable. In addition to therapeutic applications, the combined expression profiles of several miRNAs mentioned can also be used as a diagnostic marker. Despite the importance of miR-218 in gynecological cancers based on a combination of studies, there is no study on the lncRNAs that target this miRNA in gynecological cancers, so it seems that further studies in this area could be very valuable. There is a column in Table 10 that presents a list of lncRNAs that target miRNAs, which can be effective for deep insight into the ceRNA network. After reviewing ncRNA studies in gynecological cancers, it was found that genes include TEN, ZEB1, ZEB2, HMGA2, MACC1, TIMP2, TWIST1, MMP-9, Tiam1, EGFR, LVSI, NOB1, and mTOR have been studied as the most important genes involved in gynecological cancers. These data are sorted in Table 11 based on the number of studies, in addition to their targeting miRNAs. PTEN, for example, is one of the most well-known tumor suppressors, and ZEB1 and ZEB2, the most important genes involved in EMT, are at the top of the table. In order to introduce and identify miRNAs with study potential in research, Table 12 was created and based on it, Van 1 diagram was drawn. Among the miRNAs examined, only 22 miRNAs were screened in all three gynecological cancers. In addition, there are over 50 miRNAs on the list that have been studied in only two of the three cancers and have the potential for research.
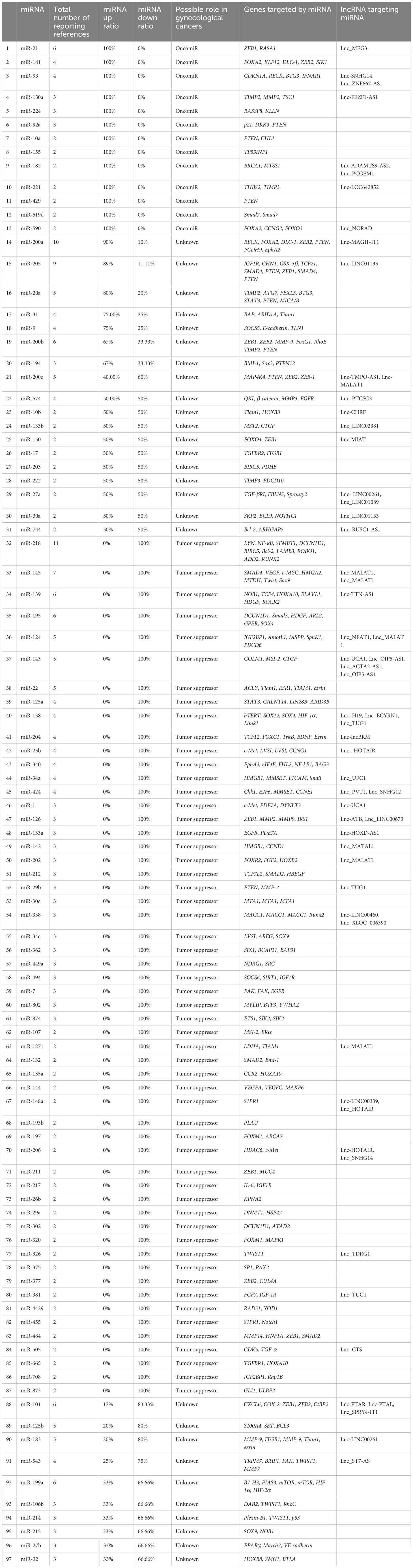
Table 10 The up or down ratio of miRNAs and their targets and lncRNA-targeting miRNA with more than one reference in metastatic gynecological cancers.
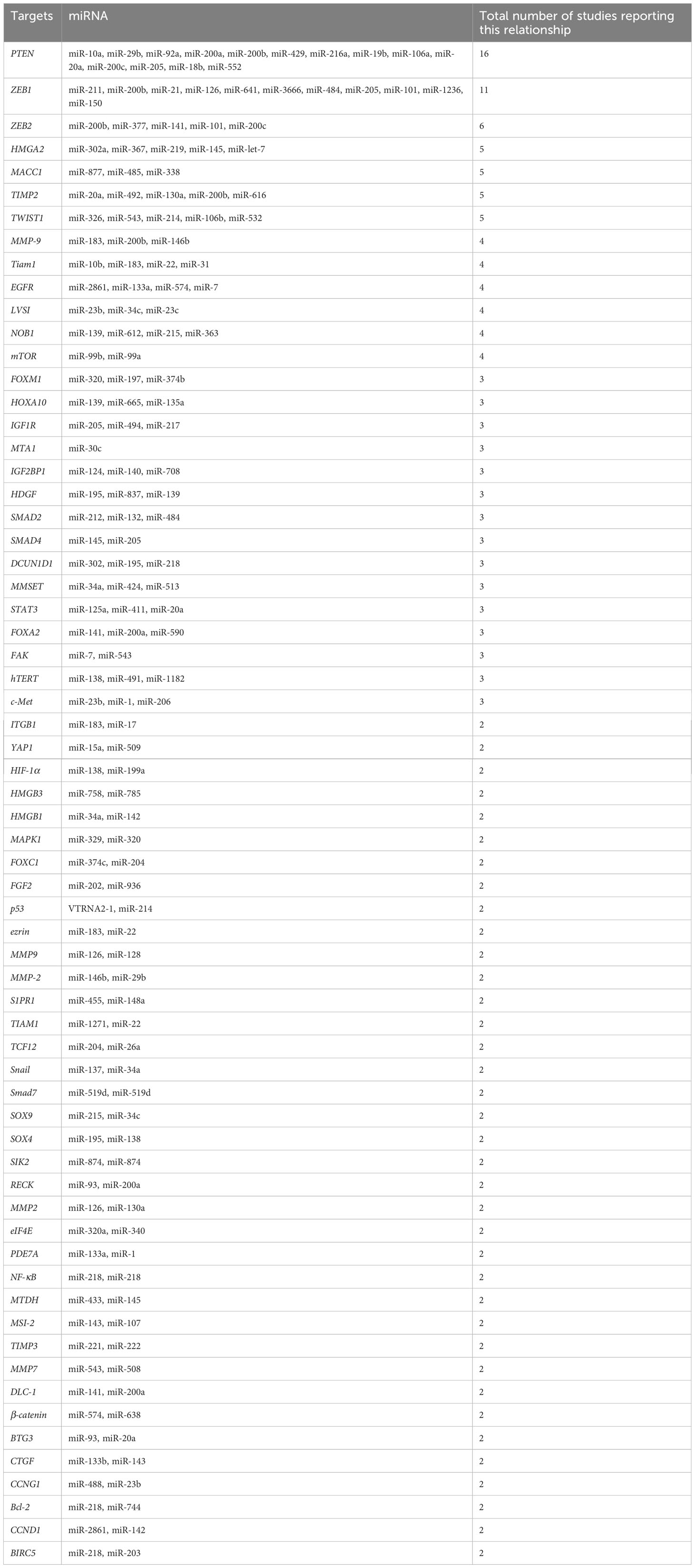
Table 11 The most important genes based on the number of studies performed and the miRNAs that target them.
Author contributions
MD, MMT, and AJ involved in conception, design, statistical analysis and drafting of the manuscript. AR, SA, SAG, SSTZ, MRH, AR, and ARA contributed in involved in the conception, interpretation of data, drafting and critically revised manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
MH declares the following potential conflicts of interest—Scientific Advisory Boards: Transdermal Cap Inc., Cleveland, OH; BeWell Global Inc., Wan Chai, Hong Kong; Hologenix Inc., Santa Monica, CA; LumiTheraInc, Poulsbo, WA; Vielight, Toronto, Canada; Bright Photomedicine, Sao Paulo, Brazil; Quantum Dynamics LLC, Cambridge, MA; Global Photon Inc., Bee Cave, TX; Medical Coherence, Boston, MA; NeuroThera, Newark, DE; JOOVV Inc., Minneapolis-St. Paul MN; AIRx Medical, Pleasanton, CA; FIR Industries, Inc., Ramsey, NJ; UVLRx Therapeutics, Oldsmar, FL; Ultralux UV Inc., Lansing, MI; Illumiheal&Petthera, Shoreline, WA; MB Lasertherapy, Houston, TX; ARRC LED, San Clemente, CA; Varuna Biomedical Corp., Incline Village, NV; Niraxx Light Therapeutics, Inc., Boston, MA; Consulting; Lexington Int., Boca Raton, FL; USHIO Corp, Japan; Merck KGaA, Darmstadt, Germany; Philips Electronics Nederland B.V. Eindhoven, Netherlands; Johnson & Johnson Inc., Philadelphia, PA; Sanofi-Aventis Deutschland GmbH, Frankfurt am Main, Germany. Stockholdings: Global Photon Inc., Bee Cave, TX; Mitonix, Newark, DE.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Wartko P, Sherman ME, Yang HP, Felix AS, Brinton LA, Trabert B. Recent changes in endometrial cancer trends among menopausal-age US women. Cancer Epidemiol (2013) 37(4):374–7. doi: 10.1016/j.canep.2013.03.008
3. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell (2011) 147(2):275–92. doi: 10.1016/j.cell.2011.09.024
4. Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin Cancer Biol (2015) 35 Suppl:S185–s198. doi: 10.1016/j.semcancer.2015.03.004
5. Zhang Y, Weinberg RA. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front Med (2018) 12(4):361–73. doi: 10.1007/s11684-018-0656-6
6. Tamtaji OR, Derakhshan M, Rashidi Noshabad FZ, Razaviyan J, Hadavi R, Jafarpour H, et al. Non-coding RNAs and brain tumors: insights into their roles in apoptosis. Front Cell Dev Biol (2022) 9:792185. doi: 10.3389/fcell.2021.792185
7. Rajabi A, Kayedi M, Rahimi S, Dashti F, Mirazimi SMA, Homayoonfal M, et al. Non-coding RNAs and glioma: Focus on cancer stem cells. Mol Therapy-Oncolytics (2022). doi: 10.1016/j.omto.2022.09.005
8. Wang Y, Mo Y, Yang X, Zhou R, Wu Z, He Y, et al. Long non-coding RNA AFAP1-AS1 is a novel biomarker in various cancers: a systematic review and meta-analysis based on the literature and GEO datasets. Oncotarget (2017) 8(60):102346–60. doi: 10.18632/oncotarget.21830
9. Wei F, Wu Y, Tang L, He Y, Shi L, Xiong F, et al. BPIFB1 (LPLUNC1) inhibits migration and invasion of nasopharyngeal carcinoma by interacting with VTN and VIM. Br J Cancer (2018) 118(2):233–47. doi: 10.1038/bjc.2017.385
10. Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer (2021) 21(7):446–60. doi: 10.1038/s41568-021-00353-1
11. Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S, et al. Circular RNAs in human cancer. Mol Cancer (2017) 16(1):25. doi: 10.1186/s12943-016-0574-7
12. He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res (2017) 36(1):145. doi: 10.1186/s13046-017-0614-1
13. Moreno-Smith M, Lutgendorf SK, Sood AK. Impact of stress on cancer metastasis. Future Oncol (2010) 6(12):1863–81. doi: 10.2217/fon.10.142
14. Voronov E, Shouval DS, Krelin Y, Cagnano E, Benharroch D, Iwakura Y, et al. IL-1 is required for tumor invasiveness and angiogenesis. Proc Natl Acad Sci (2003) 100(5):2645–50. doi: 10.1073/pnas.0437939100
15. Chan JK, Hamilton CA, Cheung MK, Karimi M, Baker J, Gall JM, et al. Enhanced killing of primary ovarian cancer by retargeting autologous cytokine-induced killer cells with bispecific antibodies: a preclinical study. Clin Cancer Res (2006) 12(6):1859–67. doi: 10.1158/1078-0432.CCR-05-2019
16. Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol (2002) 3(11):991–8. doi: 10.1038/ni1102-991
17. Paget S. The distribution of secondary growths in cancer of the breast. Cancer Metastasis Rev (1989) 8:98–101.
18. Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduction Targeted Ther (2020) 5(1):1–17. doi: 10.1038/s41392-020-0134-x
19. Clark AG, Vignjevic DM. Modes of cancer cell invasion and the role of the microenvironment. Curr Opin Cell Biol (2015) 36:13–22. doi: 10.1016/j.ceb.2015.06.004
20. Cheung KJ, Ewald AJ. A collective route to metastasis: Seeding by tumor cell clusters. Science (2016) 352(6282):167–9. doi: 10.1126/science.aaf6546
21. Lai X, Li Q, Wu F, Lin J, Chen J, Zheng H, et al. Epithelial-mesenchymal transition and metabolic switching in cancer: Lessons from somatic cell reprogramming. Front Cell Dev Biol (2020) 760. doi: 10.3389/fcell.2020.00760
23. Hamidi H, Ivaska J. Every step of the way: integrins in cancer progression and metastasis. Nat Rev Cancer (2018) 18(9):533–48. doi: 10.1038/s41568-018-0038-z
24. Jayatilaka H, Tyle P, Chen JJ, Kwak M, Ju J, Kim HJ, et al. Synergistic IL-6 and IL-8 paracrine signalling pathway infers a strategy to inhibit tumour cell migration. Nat Commun (2017) 8(1):1–12. doi: 10.1038/ncomms15584
25. Boire A, Zou Y, Shieh J, Macalinao DG, Pentsova E, Massagué J. Complement component 3 adapts the cerebrospinal fluid for leptomeningeal metastasis. Cell (2017) 168(6):1101–1113. e13. doi: 10.1016/j.cell.2017.02.025
26. Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer (2017) 17(5):302–17. doi: 10.1038/nrc.2017.6
27. Aziz SW, Aziz MH. Chapter 5 - cervical cancer metastasis. In: Ahmad A, editor. Introduction to cancer metastasis. Academic Press (2017). p. 77–94.
28. Fidler IJ. The organ microenvironment and cancer metastasis. Differentiation (2002) 70(9-10):498–505. doi: 10.1046/j.1432-0436.2002.700904.x
29. Chen C, Ge X, Zhao Y, Wang D, Ling L, Zheng S, et al. Molecular alterations in metastatic ovarian cancer from gastrointestinal cancer. Front Oncol (2020) 2713. doi: 10.3389/fonc.2020.605349
30. Pitluk H, Poticha S. Carcinoma of the colon and rectum in patients less than 40 years of age. Surgery Gynecology Obstetrics (1983) 157(4):335–7.
31. Hanahan D. Signaling vascular morphogenesis and maintenance. Science (1997) 277(5322):48–50. doi: 10.1126/science.277.5322.48
32. Tan DS, Agarwal R, Kaye SB. Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol (2006) 7(11):925–34. doi: 10.1016/S1470-2045(06)70939-1
33. Stouffer RL, Martı́nez-Chequer JC, Molskness TA, Xu F, Hazzard TM. Regulation and action of angiogenic factors in the primate ovary. Arch Med Res (2001) 32(6):567–75. doi: 10.1016/S0188-4409(01)00323-X
34. Wulff C, Wilson H, Largue P, Duncan WC, Armstrong DG, Fraser HM. Angiogenesis in the human corpus luteum: localization and changes in angiopoietins, tie-2, and vascular endothelial growth factor messenger ribonucleic acid. J Clin Endocrinol Metab (2000) 85(11):4302–9.
35. Christenson LK, Stouffer RL. Follicle-stimulating hormone and luteinizing hormone/chorionic gonadotropin stimulation of vascular endothelial growth factor production by macaque granulosa cells from pre-and periovulatory follicles. J Clin Endocrinol Metab (1997) 82(7):2135–42. doi: 10.1210/jc.82.7.2135
36. LeCouter J, Kowalski J, Foster J, Hass P, Zhang Z, Dillard-Telm L, et al. Identification of an angiogenic mitogen selective for endocrine gland endothelium. Nature (2001) 412(6850):877–84. doi: 10.1038/35091000
37. Gupta RA, Tejada LV, Tong BJ, Das SK, Morrow JD, Dey SK, et al. Cyclooxygenase-1 is overexpressed and promotes angiogenic growth factor production in ovarian cancer. Cancer Res (2003) 63(5):906–11.
38. Parrott JA, Skinner MK. Expression and action of hepatocyte growth factor in human and bovine normal ovarian surface epithelium and ovarian cancer. Biol Reprod (2000) 62(3):491–500. doi: 10.1095/biolreprod62.3.491
40. Buys SS, Partridge E, Black A, Johnson CC, Lamerato L, Isaacs C, et al. Effect of screening on ovarian cancer mortality: the prostate, lung, colorectal and ovarian (PLCO) cancer screening randomized controlled trial. Jama (2011) 305(22):2295–303. doi: 10.1001/jama.2011.766
41. Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res (2007) 67(18):8699–707. doi: 10.1158/0008-5472.CAN-07-1936
42. Fu X, Li Y, Alvero A, Li J, Wu Q, Xiao Q, et al. MicroRNA-222-3p/GNAI2/AKT axis inhibits epithelial ovarian cancer cell growth and associates with good overall survival. Oncotarget (2016) 7(49):80633–54. doi: 10.18632/oncotarget.13017
43. Wu H, Xiao Z, Zhang H, Wang K, Liu W, Hao Q. MiR-489 modulates cisplatin resistance in human ovarian cancer cells by targeting Akt3. Anti-Cancer Drugs (2014) 25(7):799–809. doi: 10.1097/CAD.0000000000000107
44. Hua Y, Larsen N, Kalyana-Sundaram S, Kjems J, Chinnaiyan AM, Peter ME. miRConnect 2.0: identification of oncogenic, antagonistic miRNA families in three human cancers. BMC Genomics (2013) 14:179. doi: 10.1186/1471-2164-14-179
45. Zhou Z, Rawnsley DR, Goddard LM, Pan W, Cao XJ, Jakus Z, et al. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev Cell (2015) 32(2):168–80. doi: 10.1016/j.devcel.2014.12.009
46. Chen PY, Chang WS, Lai YK, Wu CW. C-myc regulates the coordinated transcription of brain disease-related PDCD10-SERPINI1 bidirectional gene pair. Mol Cell Neurosci (2009) 42(1):23–32. doi: 10.1016/j.mcn.2009.05.001
47. DiStefano PV, Kuebel JM, Sarelius IH, Glading AJ. KRIT1 protein depletion modifies endothelial cell behavior via increased vascular endothelial growth factor (VEGF) signaling. J Biol Chem (2014) 289(47):33054–65. doi: 10.1074/jbc.M114.582304
48. Chen PY, Chang WS, Chou RH, Lai YK, Lin SC, Chi CY, et al. Two non-homologous brain diseases-related genes, SERPINI1 and PDCD10, are tightly linked by an asymmetric bidirectional promoter in an evolutionarily conserved manner. BMC Mol Biol (2007) 8:2. doi: 10.1186/1471-2199-8-2
49. Zhang M, Dong L, Shi Z, Jiao S, Zhang Z, Zhang W, et al. Structural mechanism of CCM3 heterodimerization with GCKIII kinases. Structure (2013) 21(4):680–8. doi: 10.1016/j.str.2013.02.015
50. Fan L, Lei H, Zhang S, Peng Y, Fu C, Shu G, et al. Non-canonical signaling pathway of SNAI2 induces EMT in ovarian cancer cells by suppressing miR-222-3p transcription and upregulating PDCD10. Theranostics (2020) 10(13):5895. doi: 10.7150/thno.43198
51. Liu L, Ning Y, Yi J, Yuan J, Fang W, Lin Z, et al. miR-6089/MYH9/β-catenin/c-Jun negative feedback loop inhibits ovarian cancer carcinogenesis and progression. Biomedicine Pharmacotherapy (2020) 125:109865. doi: 10.1016/j.biopha.2020.109865
52. Soni M, Patel Y, Markoutsa E, Jie C, Liu S, Xu P, et al. Autophagy, cell viability, and chemoresistance are regulated by miR-489 in breast cancer. Mol Cancer Res (2018) 16(9):1348–60. doi: 10.1158/1541-7786.MCR-17-0634
53. Schüler-Toprak S, Moehle C, Skrzypczak M, Ortmann O, Treeck O. Effect of estrogen receptor β agonists on proliferation and gene expression of ovarian cancer cells. BMC Cancer (2017) 17:1–9.
54. Cesa LC, Shao H, Srinivasan SR, Tse E, Jain C, Zuiderweg ER, et al. X-linked inhibitor of apoptosis protein (XIAP) is a client of heat shock protein 70 (Hsp70) and a biomarker of its inhibition. J Biol Chem (2018) 293(7):2370–80. doi: 10.1074/jbc.RA117.000634
55. Ayachi O, Barlin M, Broxtermann PN, Kashkar H, Mauch C, Zigrino P. The x-linked inhibitor of apoptosis protein (XIAP) is involved in melanoma invasion by regulating cell migration and survival. Cell Oncol (2019) 42(3):319–29. doi: 10.1007/s13402-019-00427-1
56. Jiang H, Li L, Jiang P, Wang Y. MicroRNA-489 targets XIAP to inhibit the biological progression of ovarian cancer via regulating PI3K/Akt signaling pathway and epithelial-to-mesenchymal transition. Eur Rev Med Pharmacol Sci (2020) 24:4113–22.
57. Liu J, Cao L, Zhao N, Feng Y, Yu Z, Li Y, et al. miR−338−3p inhibits A549 lung cancer cell proliferation and invasion by targeting AKT and β−catenin signaling pathways. Mol Med Rep (2019) 20(1):33–40.
58. Zhang Y, Shi B, Chen J, Hu L, Zhao C. MiR-338-3p targets pyruvate kinase M2 and affects cell proliferation and metabolism of ovarian cancer. Am J Trans Res (2016) 8(7):3266.
59. Zhang R, Shi H, Ren F, Liu Z, Ji P, Zhang W, et al. Down-regulation of miR-338-3p and up-regulation of MACC1 indicated poor prognosis of epithelial ovarian cancer patients. J Cancer (2019) 10(6):1385. doi: 10.7150/jca.29502
60. Zhang R, Shi H, Ren F, Feng W, Cao Y, Li G, et al. MicroRNA-338-3p suppresses ovarian cancer cells growth and metastasis: implication of wnt/catenin beta and MEK/ERK signaling pathways. J Exp Clin Cancer Res (2019) 38(1):1–13. doi: 10.1186/s13046-019-1494-3
61. Zhou X, Tao H. Overexpression of microRNA−936 suppresses non−small cell lung cancer cell proliferation and invasion via targeting E2F2. Exp Ther Med (2018) 16(3):2696–702.
62. Li C, Yu S, Wu S, Ni Y, Pan Z. MicroRNA-936 targets FGF2 to inhibit epithelial ovarian cancer aggressiveness by deactivating the PI3K/Akt pathway. OncoTargets Ther (2019) 12:5311. doi: 10.2147/OTT.S213231
63. Lu X, Han Y, Han Y, Huang M, You J, Liu Y, et al. MicroRNA-650 suppresses KLF12 expression to regulate growth and metastasis of human ovarian cancer cells. Acta Biochim Pol (2022) 69(4):745–51. doi: 10.18388/abp.2020_5987
64. Shan L, Song P, Zhao Y, An N, Xia Y, Qi Y, et al. miR-600 promotes ovarian cancer cells stemness, proliferation and metastasis via targeting KLF9. J Ovarian Res (2022) 15(1):52. doi: 10.1186/s13048-022-00981-7
65. Xiong T, Wang Y, Zhang Y, Yuan J, Zhu C, Jiang W. lncRNA AC005224.4/miR-140-3p/SNAI2 regulating axis facilitates the invasion and metastasis of ovarian cancer through epithelial-mesenchymal transition. Chin Med J (Engl) (2023) 136(9):1098–110.
66. Xue F, Li QR, Xu YH, Zhou HB. MicroRNA-139-3p inhibits the growth and metastasis of ovarian cancer by inhibiting ELAVL1. OncoTargets Ther (2019) 12:8935. doi: 10.2147/OTT.S210739
67. Guo J, Wang X, Sun L. MicroRNA-488 inhibits ovarian cancer cell metastasis through regulating CCNG1 and p53 expression. Eur Rev Med Pharmacol Sci (2020) 24(6):2902–10.
68. Srivastava AK, Banerjee A, Cui T, Han C, Cai S, Liu L, et al. Inhibition of miR-328–3p impairs cancer stem cell function and prevents metastasis in ovarian cancer. Cancer Res (2019) 79(9):2314–26. doi: 10.1158/0008-5472.CAN-18-3668
69. Huang Z, Li Q, Luo K, Zhang Q, Geng J, Zhou X, et al. miR-340-FHL2 axis inhibits cell growth and metastasis in ovarian cancer. Cell Death Dis (2019) 10(5):1–16. doi: 10.1038/s41419-019-1604-3
70. Buranjiang G, Kuerban R, Abuduwanke A, Li X, Kuerban G. MicroRNA-331-3p inhibits proliferation and metastasis of ovarian cancer by targeting RCC2. Arch Med Science: AMS (2019) 15(6):1520. doi: 10.5114/aoms.2018.77858
71. Wang L, Zhao S, Yu M. Mechanism of low expression of miR-30a-5p on epithelial–mesenchymal transition and metastasis in ovarian cancer. DNA Cell Biol (2019) 38(4):341–51. doi: 10.1089/dna.2018.4396
72. Zhu F, Li J, Wang L. MicroRNA-1-3p inhibits the growth and metastasis of ovarian cancer cells by targeting DYNLT3. Eur Rev Med Pharmacol Sci (2020) 24(17):8713–21.
73. Yang S, Yang R, Lin R, Si L. MicroRNA-375 inhibits the growth, drug sensitivity and metastasis of human ovarian cancer cells by targeting PAX2. J BUON (2019) 24(6):2341–6.
74. Xing F, Wang S, Zhou J. The expression of microRNA-598 inhibits ovarian cancer cell proliferation and metastasis by targeting URI. Mol Therapy-Oncolytics (2019) 12:9–15. doi: 10.1016/j.omto.2018.12.002
75. Zeng S, Liu S, Feng J, Gao J, Xue F. MicroRNA−32 promotes ovarian cancer cell proliferation and motility by targeting SMG1. Oncol Lett (2020) 20(1):733–41. doi: 10.3892/ol.2020.11624
76. Fan B, Chen L, Yuan Y, Xiao H, Lv X, Xia Z. MiR-15a-3p suppresses the growth and metastasis of ovarian cancer cell by targeting Twist1. Eur Rev Med Pharmacol Sci (2019) 23(5):1934–46.
77. Mei J, Huang Y, Hao L, Liu Y, Yan T, Qiu T, et al. DAAM1-mediated migration and invasion of ovarian cancer cells are suppressed by miR-208a-5p. Pathology-Research Pract (2019) 215(7):152452. doi: 10.1016/j.prp.2019.152452
78. Bi Y-N, Guan J-P, Wang L, Li P, Yang F-X. Clinical significance of microRNA-125b and its contribution to ovarian carcinogenesis. Bioengineered (2020) 11(1):939–48. doi: 10.1080/21655979.2020.1814660
79. Park GB, Kim D. MicroRNA-503-5p inhibits the CD97-mediated JAK2/STAT3 pathway in metastatic or paclitaxel-resistant ovarian cancer cells. Neoplasia (2019) 21(2):206–15. doi: 10.1016/j.neo.2018.12.005
80. Barbier J, Chen X, Sanchez G, Cai M, Helsmoortel M, Higuchi T, et al. An NF90/NF110-mediated feedback amplification loop regulates dicer expression and controls ovarian carcinoma progression. Cell Res (2018) 28(5):556–71. doi: 10.1038/s41422-018-0016-8
81. Gao S, Bian T, Su M, Liu Y, Zhang Y. miR−26a inhibits ovarian cancer cell proliferation, migration and invasion by targeting TCF12. Oncol Rep (2020) 43(1):368–74.
82. Ge T, Liu T, Guo L, Chen Z, Lou G. MicroRNA-302 represses epithelial-mesenchymal transition and cisplatin resistance by regulating ATAD2 in ovarian carcinoma. Exp Cell Res (2020) 396(1):112241. doi: 10.1016/j.yexcr.2020.112241
83. Zheng Y, Yang X, Wang C, Zhang S, Wang Z, Li M, et al. HDAC6, modulated by miR-206, promotes endometrial cancer progression through the PTEN/AKT/mTOR pathway. Sci Rep (2020) 10(1):1–12. doi: 10.1038/s41598-020-60271-4
84. Lin M, Xia B, Qin L, Chen H, Lou G. S100A7 regulates ovarian cancer cell metastasis and chemoresistance through MAPK signaling and is targeted by miR-330-5p. DNA Cell Biol (2018) 37(5):491–500. doi: 10.1089/dna.2017.3953
85. Zhuang R-J, Bai X-X, Liu W. MicroRNA-23a depletion promotes apoptosis of ovarian cancer stem cell and inhibits cell migration by targeting DLG2. Cancer Biol Ther (2019) 20(6):897–911. doi: 10.1080/15384047.2019.1579960
86. Zhou J, Zhang X, Li W, Chen Y. MicroRNA-145-5p regulates the proliferation of epithelial ovarian cancer cells via targeting SMAD4. J Ovarian Res (2020) 13:1–9. doi: 10.1186/s13048-020-00656-1
87. Yang B, Sun L, Liang L. MiRNA-802 suppresses proliferation and migration of epithelial ovarian cancer cells by targeting YWHAZ. J Ovarian Res (2019) 12(1):1–8. doi: 10.1186/s13048-019-0576-3
88. Li R, Wu H, Jiang H, Wang Q, Dou Z, Ma H, et al. FBLN5 is targeted by microRNA−27a−3p and suppresses tumorigenesis and progression in high−grade serous ovarian carcinoma. Oncol Rep (2020) 44(5):2143–51.
89. Liu DT, Yao HR, Li YY, Song YY, Su MY. MicroRNA−19b promotes the migration and invasion of ovarian cancer cells by inhibiting the PTEN/AKT signaling pathway. Oncol Lett (2018) 16(1):559–65.
90. Wang B, Li X, Zhao G, Yan H, Dong P, Watari H, et al. miR-203 inhibits ovarian tumor metastasis by targeting BIRC5 and attenuating the TGFβ pathway. J Exp Clin Cancer Res (2018) 37(1):1–9.
91. Yu H, Pan S. MiR-202-5p suppressed cell proliferation, migration and invasion in ovarian cancer via regulating HOXB2. Eur Rev Med Pharmacol Sci (2020) 24:2256–63.
92. Chu P, Liang A, Jiang A, Zong L. miR-205 regulates the proliferation and invasion of ovarian cancer cells via suppressing PTEN/SMAD4 expression. Oncol Lett (2018) 2018:7571–8. doi: 10.3892/ol.2018.8313
93. Garrido MP, Torres I, Avila A, Chnaiderman J, Valenzuela-Valderrama M, Aramburo J, et al. NGF/TRKA decrease miR-145-5p levels in epithelial ovarian cancer cells. Int J Mol Sci (2020) 21(20):7657. doi: 10.3390/ijms21207657
94. Wahab NA, Othman Z, Nasri NWM, Mokhtar MH, Ibrahim SF, Hamid AA, et al. Inhibition of miR-141 and miR-200a increase DLC-1 and ZEB2 expression, enhance migration and invasion in metastatic serous ovarian cancer. Int J Environ Res Public Health (2020) 17(8):2766. doi: 10.3390/ijerph17082766
95. Liu J, Li C, Jiang Y, Wan Y, Zhou S, Cheng W. Tumor-suppressor role of miR-139-5p in endometrial cancer. Cancer Cell Int (2018) 18(1):1–9. doi: 10.1186/s12935-018-0545-8
96. Salem M, Shan Y, Bernaudo S, Peng C. miR-590-3p targets cyclin G2 and FOXO3 to promote ovarian cancer cell proliferation, invasion, and spheroid formation. Int J Mol Sci (2019) 20(8):1810. doi: 10.3390/ijms20081810
97. Zheng J, Zhou Y, Li X, Hu J. MiR-574-3p exerts as a tumor suppressor in ovarian cancer through inhibiting MMP3 expression. Eur Rev Med Pharmacol Sci (2019) 23(16):6839–48.
98. Shi C, Yang Y, Zhang L, Yu J, Qin S, Xu H, et al. MiR-200a-3p promoted the malignant behaviors of ovarian cancer cells through regulating PCDH9. OncoTargets Ther (2019) 12:8329. doi: 10.2147/OTT.S220339
99. Liu J, Gu Z, Tang Y, Hao J, Zhang C, Yang X. Tumour-suppressive microRNA-424-5p directly targets CCNE1 as potential prognostic markers in epithelial ovarian cancer. Cell Cycle (2018) 17(3):309–18. doi: 10.1080/15384101.2017.1407894
100. Zha J, Chen D. MiR-655-3p inhibited proliferation and migration of ovarian cancer cells by targeting RAB1A. Eur Rev Med Pharmacol Sci (2019) 23(9):3627–34.
101. Jiang B, Zhu S-J, Xiao S-S, Xue M. MiR-217 inhibits M2-like macrophage polarization by suppressing secretion of interleukin-6 in ovarian cancer. Inflammation (2019) 42(5):1517–29. doi: 10.1007/s10753-019-01004-2
102. Xiao S, Li Y, Pan Q, Ye M, He S, Tian Q, et al. MiR-34c/SOX9 axis regulates the chemoresistance of ovarian cancer cell to cisplatin-based chemotherapy. J Cell Biochem (2019) 120(3):2940–53. doi: 10.1002/jcb.26865
103. Li J, Shao W, Zhao J. MiR-520a-3p inhibits malignant progression of epithelial ovarian cancer by targeting SUV39H1 expression. Hum Cell (2020) 2020:1–9.
104. Guo F, Zhang K, Li M, Cui L, Liu G, Yan Y, et al. miR−508−3p suppresses the development of ovarian carcinoma by targeting CCNA2 and MMP7. Int J Oncol (2020) 57(1):264–76. doi: 10.3892/ijo.2020.5055
105. Liu F, Zhang G, Lv S, Wen X, Liu P. miRNA-301b-3p accelerates migration and invasion of high-grade ovarian serous tumor via targeting CPEB3/EGFR axis. J Cell Biochem (2019) 120(8):12618–27. doi: 10.1002/jcb.28528
106. Yang L, Ma H. MiRNA-584 suppresses the progression of ovarian cancer by negatively regulating LPIN1. Eur Rev Med Pharmacol Sci (2020) 24:1062–71.
107. Zhu Y, Chen P, Shi L, Zhu T, Chen X. MiR-4429 suppresses the malignant development of ovarian cancer by targeting YOD1. Eur Rev Med Pharmacol Sci (2020) 24(17):8722–30.
108. Xia B, Lin M, Dong W, Chen H, Li B, Zhang X, et al. Upregulation of miR-874-3p and miR-874-5p inhibits epithelial ovarian cancer malignancy via SIK2. J Biochem Mol Toxicol (2018) 32(8):e22168. doi: 10.1002/jbt.22168
109. Wei H, Tang Q, Zhang K, Sun J, Ding R. miR-532-5p is a prognostic marker and suppresses cells proliferation and invasion by targeting TWIST1 in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci (2018) 22(18):5842–50.
110. Zhang X, Sun B, Tian S, Li L, Zhao Y, Shi P. MicroRNA-132 reverses cisplatin resistance and metastasis in ovarian cancer by the targeted regulation on bmi-1. Eur Rev Med Pharmacol Sci (2019) 23(9):3635–44.
111. Zhao W, Han T, Li B, Ma Q, Yang P, Li H. miR-552 promotes ovarian cancer progression by regulating PTEN pathway. J Ovarian Res (2019) 12(1):1–10. doi: 10.1186/s13048-019-0589-y
112. Zeng J, Li YK, Quan FF, Zeng X, Chen CY, Zeng T, et al. Propofol−induced miR−125a−5p inhibits the proliferation and metastasis of ovarian cancer by suppressing LIN28B. Mol Med Rep (2020) 22(2):1507–17. doi: 10.3892/mmr.2020.11223
113. Setiawan VW, Yang HP, Pike MC, McCann SE, Yu H, Xiang YB, et al. And II endometrial cancers: have they different risk factors? J Clin Oncol (2013) 31(20):2607–18. doi: 10.1200/JCO.2012.48.2596
114. Gottwald L, Pluta P, Piekarski J, Spych M, Hendzel K, Topczewska-Tylinska K, et al. Long-term survival of endometrioid endometrial cancer patients. Arch Med Sci (2010) 6(6):937–44. doi: 10.5114/aoms.2010.19305
115. Tarone RE, Chu KC. Age-period-cohort analyses of breast-, ovarian-, endometrial- and cervical-cancer mortality rates for caucasian women in the USA. J Epidemiol Biostat (2000) 5(4):221–31.
116. Kasius JC, Pijnenborg J, Lindemann K, Forsse D, van Zwol J, Kristensen GB, et al. Risk stratification of endometrial cancer patients: FIGO stage, biomarkers and molecular classification. Cancers (2021) 13(22):5848. doi: 10.3390/cancers13225848
117. Liu F, Zhao X, Qian Y, Zhang J, Zhang Y, Yin R. MiR-206 inhibits head and neck squamous cell carcinoma cell progression by targeting HDAC6 via PTEN/AKT/mTOR pathway. Biomedicine Pharmacotherapy (2017) 96:229–37. doi: 10.1016/j.biopha.2017.08.145
118. Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer (2006) 6(1):38–51. doi: 10.1038/nrc1779
119. Bazzaro M, Lin Z, Santillan A, Lee MK, Wang M-C, Chan KC, et al. Ubiquitin proteasome system stress underlies synergistic killing of ovarian cancer cells by bortezomib and a novel HDAC6 inhibitor. Clin Cancer Res (2008) 14(22):7340–7. doi: 10.1158/1078-0432.CCR-08-0642
120. Ma J, Li D, Kong F-F, Yang D, Yang H, Ma X-X. miR-302a-5p/367-3p-HMGA2 axis regulates malignant processes during endometrial cancer development. J Exp Clin Cancer Res (2018) 37(1):1–17. doi: 10.1186/s13046-018-0686-6
121. Shi S, Chen X, Liu H, Yu K, Bao Y, Chai J, et al. LGR5 acts as a target of miR-340-5p in the suppression of cell progression and drug resistance in breast cancer via wnt/β-catenin pathway. Gene (2019) 683:47–53. doi: 10.1016/j.gene.2018.10.014
122. Zhang HH, Li R, Li YJ, Yu XX, Sun QN, Li AY, et al. eIF4E−related miR−320a and miR−340−5p inhibit endometrial carcinoma cell metastatic capability by preventing TGF−β1−induced epithelial−mesenchymal transition. Oncol Rep (2020) 43(2):447–60.
123. Hu J, Zhang L, Mei Z, Jiang Y, Yi Y, Liu L, et al. Interaction of E3 ubiquitin ligase MARCH7 with long noncoding RNA MALAT1 and autophagy-related protein ATG7 promotes autophagy and invasion in ovarian cancer. Cell Physiol Biochem (2018) 47(2):654–66. doi: 10.1159/000490020
124. Zhao B, Ito K, Iyengar PV, Hirose S, Nakamura N. MARCH7 E3 ubiquitin ligase is highly expressed in developing spermatids of rats and its possible involvement in head and tail formation. Histochem Cell Biol (2013) 139(3):447–60. doi: 10.1007/s00418-012-1043-z
125. Szigyarto CA, Sibbons P, Williams G, Uhlen M, Metcalfe SM. The E3 ligase axotrophin/MARCH-7: protein expression profiling of human tissues reveals links to adult stem cells. J Histochem Cytochem (2010) 58(4):301–8. doi: 10.1369/jhc.2009.954420
126. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet (2004) 5(7):522–31. doi: 10.1038/nrg1379
127. Chen D, Si W, Shen J, Du C, Lou W, Bao C, et al. miR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis (2018 2018) 9(2):188. doi: 10.1038/s41419-017-0211-4
128. Liu L, Hu J, Yu T, You S, Zhang Y, Hu L. miR-27b-3p/MARCH7 regulates invasion and metastasis of endometrial cancer cells through snail-mediated pathway. Acta Biochim Biophys Sin (2019) 51(5):492–500. doi: 10.1093/abbs/gmz030
129. Onate SA, Tsai SY, Tsai M-J, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science (1995) 270(5240):1354–7. doi: 10.1126/science.270.5240.1354
130. Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SA, et al. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Institute (2003) 95(5):353–61. doi: 10.1093/jnci/95.5.353
131. Kershah SM, Desouki MM, Koterba KL, Rowan BG. Expression of estrogen receptor coregulators in normal and malignant human endometrium. Gynecologic Oncol (2004) 92(1):304–13. doi: 10.1016/j.ygyno.2003.10.007
132. Sakaguchi H, Fujimoto J, Sun W-S, Tamaya T. Clinical implications of steroid receptor coactivator (SRC)-3 in uterine endometrial cancers. J Steroid Biochem Mol Biol (2007) 104(3-5):237–40. doi: 10.1016/j.jsbmb.2007.03.007
133. Hu Y, Wu A-Y, Xu C, Song K-Q, Wang W-J, Yin X, et al. MicroRNA-449a inhibits tumor metastasis through AKT/ERK1/2 inactivation by targeting steroid receptor coactivator (SRC) in endometrial cancer. J Cancer (2019) 10(2):547. doi: 10.7150/jca.27748
134. Chen C, Zhang Q, Kong B. miRNA-576-5p promotes endometrial cancer cell growth and metastasis by targeting ZBTB4. Clin Transl Oncol (2023) 25(3):706–20.
135. Sun X, Hou L, Qiu C, Kong B. MiR-501 promotes tumor proliferation and metastasis by targeting HOXD10 in endometrial cancer. Cell Mol Biol Lett (2021) 26(1):20. doi: 10.1186/s11658-021-00268-7
136. Wu AY, Hu Y, Cang W, Li D, Wang WJ, Tian Q, et al. Suppressive effect of microRNA-449a on the NDRG1/PTEN/AKT axis regulates endometrial cancer growth and metastasis. Exp Cell Res (2019) 382(2):111468. doi: 10.1016/j.yexcr.2019.06.013
137. Sun X, Dongol S, Qiu C, Xu Y, Sun C, Zhang Z, et al. miR-652 promotes tumor proliferation and metastasis by targeting RORA in endometrial cancer. Mol Cancer Res (2018) 16(12):1927–39. doi: 10.1158/1541-7786.MCR-18-0267
138. Tian Y, Chen Y, Han A. MiR-1271 inhibits cell proliferation and metastasis by targeting LDHA in endometrial cancer. Eur Rev Med Pharmacol Sci (2019) 23(13):5648–56.
139. Xu J. MicroRNA-93-5p/IFNAR1 axis accelerates metastasis of endometrial carcinoma by activating the STAT3 pathway. Eur Rev Med Pharmacol Sci (2019) 23(13):5657–66.
140. Li XC, Hai JJ, Tan YJ, Yue QF, Liu LJ. MiR-218 suppresses metastasis and invasion of endometrial cancer via negatively regulating ADD2. Eur Rev Med Pharmacol Sci (2019) 23(4):1408–17.
141. Wang Y, Zhang S. Berberine suppresses growth and metastasis of endometrial cancer cells via miR-101/COX-2. Biomedicine Pharmacotherapy (2018) 103:1287–93. doi: 10.1016/j.biopha.2018.04.161
142. Zhou Z, Xu Y-P, Wang L-J, Kong Y. miR-940 potentially promotes proliferation and metastasis of endometrial carcinoma through regulation of MRVI1. Bioscience Rep (2019) 39(6). doi: 10.1042/BSR20190077
143. Wang Q, Zhu W. MicroRNA−873 inhibits the proliferation and invasion of endometrial cancer cells by directly targeting hepatoma−derived growth factor. Exp Ther Med (2019) 18(2):1291–8.
144. Su Y, Wang J, Ma Z, Gong W, Yu L. miR-142 suppresses endometrial cancer proliferation in vitro and in vivo by targeting cyclin D1. DNA Cell Biol (2019) 38(2):144–50. doi: 10.1089/dna.2018.4441
145. Liu X, Wen J, Wang H, Wang Y. Long non-coding RNA LINC00460 promotes epithelial ovarian cancer progression by regulating microRNA-338-3p. Biomedicine Pharmacotherapy (2018) 108:1022–8. doi: 10.1016/j.biopha.2018.09.103
146. Bing L, Hong C, Li-Xin S, Wei G. MicroRNA-543 suppresses endometrial cancer oncogenicity via targeting FAK and TWIST1 expression. Arch Gynecology Obstetrics (2014) 290(3):533–41. doi: 10.1007/s00404-014-3219-3
147. Xu X, Kong X, Liu T, Zhou L, Wu J, Fu J, et al. Metastasis-associated protein 1, modulated by miR-30c, promotes endometrial cancer progression through AKT/mTOR/4E-BP1 pathway. Gynecologic Oncol (2019) 154(1):207–17. doi: 10.1016/j.ygyno.2019.04.005
148. Chen P, Xing T, Wang Q, Liu A, Liu H, Hu Y, et al. MicroRNA-202 inhibits cell migration and invasion through targeting FGF2 and inactivating wnt/β-catenin signaling in endometrial carcinoma. Bioscience Rep (2019) 39(10). doi: 10.1042/BSR20190680
149. Tu C, Wang F, Wan J. MicroRNA-381 inhibits cell proliferation and invasion in endometrial carcinoma by targeting the IGF-1R. Mol Med Rep (2018) 17(3):4090–8.
150. Fang Y-Y, Tan M-R, Zhou J, Liang L, Liu X-Y, Zhao K, et al. miR-214-3p inhibits epithelial-to-mesenchymal transition and metastasis of endometrial cancer cells by targeting TWIST1. OncoTargets Ther (2019) 12:9449. doi: 10.2147/OTT.S181037
151. Deng J, Wang W, Yu G, Ma X. MicroRNA−195 inhibits epithelial−mesenchymal transition by targeting g protein−coupled estrogen receptor 1 in endometrial carcinoma. Mol Med Rep (2019) 20(5):4023–32.
152. Huang Y, Yang N. MicroRNA-20a-5p inhibits epithelial to mesenchymal transition and invasion of endometrial cancer cells by targeting STAT3. Int J Clin Exp Pathol (2018) 11(12):5715.
153. Wang Y, Dong L, Liu Y. Targeting thyroid receptor interacting protein 6 by microrna-589-5p inhibits cell proliferation, migration, and invasion in endometrial carcinoma. Cancer Biotherapy Radiopharmaceuticals (2019) 34(8):529–36. doi: 10.1089/cbr.2018.2766
154. Bao W, Zhang Y, Li S, Fan Q, Qiu M, Wang Y, et al. miR−107−5p promotes tumor proliferation and invasion by targeting estrogen receptor−α in endometrial carcinoma. Oncol Rep (2019) 41(3):1575–85.
155. Dong P, Xiong Y, Yue J, Hanley SJ, Watari H. miR-34a, miR-424 and miR-513 inhibit MMSET expression to repress endometrial cancer cell invasion and sphere formation. Oncotarget (2018) 9(33):23253. doi: 10.18632/oncotarget.25298
156. Zhao X, Dai L, Yue Q, Wang H, Wang X, Li Y, et al. MiR-195 inhibits migration, invasion and epithelial-mesenchymal transition (EMT) of endometrial carcinoma cells by targeting SOX4. J Biosci (2019) 44(6):1–9. doi: 10.1007/s12038-019-9966-3
157. Shi W, Wang X, Ruan L, Fu J, Liu F, Qu J. MiR-200a promotes epithelial-mesenchymal transition of endometrial cancer cells by negatively regulating FOXA2 expression. Die Pharmazie-An Int J Pharm Sci (2017) 72(11):694–9.
158. Gong B, Yue Y, Wang R, Zhang Y, Jin Q, Zhou X. Overexpression of microRNA-194 suppresses the epithelial–mesenchymal transition in targeting stem cell transcription factor Sox3 in endometrial carcinoma stem cells. Tumor Biol (2017) 39(6):1010428317706217. doi: 10.1177/1010428317706217
159. Rabelo-Santos SH, Termini L, Boccardo E, Derchain S, Longatto-Filho A, Andreoli MA, et al. Strong SOD2 expression and HPV-16/18 positivity are independent events in cervical cancer. Oncotarget (2018) 9(31):21630–40. doi: 10.18632/oncotarget.24850
160. Pardini B, De Maria D, Francavilla A, Di Gaetano C, Ronco G, Naccarati A. MicroRNAs as markers of progression in cervical cancer: a systematic review. BMC Cancer (2018) 18(1):696. doi: 10.1186/s12885-018-4590-4
161. Taniguchi-Ponciano K, Ribas-Aparicio RM, Marrero-Rodríguez D, Arreola-De la Cruz H, Huerta-Padilla V, Muñoz N, et al. The KISS1 gene overexpression as a potential molecular marker for cervical cancer cells. Cancer biomark (2018) 22(4):709–19. doi: 10.3233/CBM-181215
162. Zhang Q, Lv R, Guo W, Li X. microRNA-802 inhibits cell proliferation and induces apoptosis in human cervical cancer by targeting serine/arginine-rich splicing factor 9. J Cell Biochem (2019) 120(6):10370–9. doi: 10.1002/jcb.28321
163. Ito S, Ueno A, Ueda T, Nakagawa H, Taniguchi H, Kayukawa N, et al. CNPY2 inhibits MYLIP-mediated AR protein degradation in prostate cancer cells. Oncotarget (2018) 9(25):17645–55. doi: 10.18632/oncotarget.24824
164. Zhao L, Zhao Y, He Y, Mao Y. miR-19b promotes breast cancer metastasis through targeting MYLIP and its related cell adhesion molecules. Oncotarget (2017) 8(38):64330. doi: 10.18632/oncotarget.19278
165. Ni M, Yan Q, Xue H, Du Y, Zhao S, Zhao Z. Identification of MYLIP gene and miRNA-802 involved in the growth and metastasis of cervical cancer cells. Cancer biomark (2021) 30(3):287–98. doi: 10.3233/CBM-201523
166. Ingebrigtsen VA, Boye K, Nesland JM, Nesbakken A, Flatmark K, Fodstad Ø. B7-H3 expression in colorectal cancer: associations with clinicopathological parameters and patient outcome. BMC Cancer (2014) 14(1):1–9. doi: 10.1186/1471-2407-14-602
167. Li W, Wang H, Zhang J, Zhai L, Chen W, Zhao C. miR-199a-5p regulates β1 integrin through ets-1 to suppress invasion in breast cancer. Cancer Sci (2016) 107(7):916–23. doi: 10.1111/cas.12952
168. Li SQ, Wang ZH, Mi XG, Liu L, Tan Y. MiR-199a/b-3p suppresses migration and invasion of breast cancer cells by downregulating PAK4/MEK/ERK signaling pathway. IUBMB Life (2015) 67(10):768–77. doi: 10.1002/iub.1433
169. Yang X, Feng KX, Li H, Wang L, Xia H. MicroRNA-199a inhibits cell proliferation, migration, and invasion and activates AKT/mTOR signaling pathway by targeting B7-H3 in cervical cancer. Technol Cancer Res Treat (2020) 19:1533033820942245. doi: 10.1177/1533033820942245
170. Dang E, Yang S, Song C, Jiang D, Li Z, Fan W, et al. BAP31, a newly defined cancer/testis antigen, regulates proliferation, migration, and invasion to promote cervical cancer progression. Cell Death Dis (2018) 9(8):1–15. doi: 10.1038/s41419-018-0824-2
171. Song L, Liu S, Yao H, Zhang L, Li Y, Xu D, et al. MiR-362-3p is downregulated by promoter methylation and independently predicts shorter OS of cervical squamous cell carcinoma. Biomedicine Pharmacotherapy (2019) 115:108944. doi: 10.1016/j.biopha.2019.108944
172. Christensen LL, Tobiasen H, Holm A, Schepeler T, Ostenfeld MS, Thorsen K, et al. MiRNA-362-3p induces cell cycle arrest through targeting of E2F1, USF2 and PTPN1 and is associated with recurrence of colorectal cancer. Int J Cancer (2013) 133(1):67–78. doi: 10.1002/ijc.28010
173. Kang H, Kim C, Lee H, Rho J, Seo J, Nam J, et al. Downregulation of microRNA-362-3p and microRNA-329 promotes tumor progression in human breast cancer. Cell Death Differentiation (2016) 23(3):484–95. doi: 10.1038/cdd.2015.116
174. Yang S, Sun Y, Jiang D, Wang J, Dang E, Li Z, et al. MiR-362 suppresses cervical cancer progression via directly targeting BAP31 and activating TGFβ/Smad pathway. Cancer Med (2021) 10(1):305–16. doi: 10.1002/cam4.3601
175. Liu J, Jiang J, Hui X, Wang W, Fang D, Ding L. Mir-758-5p suppresses glioblastoma proliferation, migration and invasion by targeting ZBTB20. Cell Physiol Biochem (2018) 48(5):2074–83. doi: 10.1159/000492545
176. Meng X, Zhao Y, Wang J, Gao Z, Geng Q, Liu X. Regulatory roles of miRNA-758 and matrix extracellular phosphoglycoprotein in cervical cancer. Exp Ther Med (2017) 14(4):2789–94. doi: 10.3892/etm.2017.4887
177. Wei H, Zhang JJ, Tang QL. MiR-638 inhibits cervical cancer metastasis through wnt/β-catenin signaling pathway and correlates with prognosis of cervical cancer patients. Eur Rev Med Pharmacol Sci (2017) 21(24):5587–93.
178. Bahrami A, Hasanzadeh M, ShahidSales S, Yousefi Z, Kadkhodayan S, Farazestanian M, et al. Clinical significance and prognosis value of wnt signaling pathway in cervical cancer. J Cell Biochem (2017) 118(10):3028–33. doi: 10.1002/jcb.25992
179. Song T, Hou X, Lin B. MicroRNA−758 inhibits cervical cancer cell proliferation and metastasis by targeting HMGB3 through the WNT/β−catenin signaling pathway. Oncol Lett (2019) 18(2):1786–92.
180. Li Y, Wei Y, Zhang H, Bai Y, Wang X, Li Q, et al. MicroRNA-154-5p suppresses cervical carcinoma growth and metastasis by silencing Cullin2 in vitro and in vivo. PeerJ (2023) 11:e15641. doi: 10.7717/peerj.15641
181. Hongwei L, Juan L, Xiaoying X, Zhijun F. MicroRNA-106b-5p (miR-106b-5p) suppresses the proliferation and metastasis of cervical cancer cells via down-regulating fibroblast growth factor 4 (FGF4) expression. Cytotechnology (2022) 74(4):469–78. doi: 10.1007/s10616-022-00536-0
182. Li H, An X, Fu Q. MiR-218 affects the invasion and metastasis of cervical cancer cells by inhibiting the expression of SFMBT1 and DCUNIDI. Cell Mol Biol (Noisy-le-grand) (2022) 68(2):81–6.
183. Shen W, Xie X, Liu M, Wang L. MicroRNA-101-5p inhibits the growth and metastasis of cervical cancer cell by inhibiting CXCL6. Eur Rev Med Pharmacol Sci (2019) 23(5):1957–68.
184. Liu C, Chen Y, Xie B, Li Y, Wei Y, Wang F. MicroRNA-215-3p suppresses the growth and metastasis of cervical cancer cell via targeting SOX9. Eur Rev Med Pharmacol Sci (2019) 23(13):5628–39.
185. Meng F, Ou J, Liu J, Li X, Meng Y, Yan L, et al. MicroRNA−877 is downregulated in cervical cancer and directly targets MACC1 to inhibit cell proliferation and invasion. Exp Ther Med (2019) 18(5):3650–8.
186. Wang S, Gao B, Yang H, Liu X, Wu X, Wang W. MicroRNA−432 is downregulated in cervical cancer and directly targets FN1 to inhibit cell proliferation and invasion. Oncol Lett (2019) 18(2):1475–82.
187. Feng J, Wang T. MicroRNA−873 serves a critical role in human cervical cancer proliferation and metastasis via regulating glioma−associated oncogene homolog 1. Exp Ther Med (2020) 19(2):1243–50.
188. Chen M, Liu L-X. MiR-525-5p repressed metastasis and anoikis resistance in cervical cancer via blocking UBE2C/ZEB1/2 signal axis. Digestive Dis Sci (2020) 65(8):2442–51. doi: 10.1007/s10620-019-05916-9
189. Tong R, Zhang J, Wang C, Li Q, Wang L, Ju M. Inhibition of miR-574-5p suppresses cell growth and metastasis and enhances chemosensitivity by targeting RNA binding protein QKI in cervical cancer cells. Naunyn-Schmiedeberg's Arch Pharmacol (2020) 393(6):951–66. doi: 10.1007/s00210-019-01772-6
190. Tang Y, Zhao Y, Ran J, Wang Y. MicroRNA−21 promotes cell metastasis in cervical cancer through modulating epithelial−mesenchymal transition. Oncol Lett (2020) 19(4):3289–95.
191. Sun Y, Cheng Y, Zhang Y, Han K. MicroRNA−889−3p targets FGFR2 to inhibit cervical cancer cell viability and invasion. Exp Ther Med (2019) 18(2):1440–8.
192. Wei Y, Jiao X, Zhang S, Xu Y, Li S, Kong B. MiR-9-5p could promote angiogenesis and radiosensitivity in cervical cancer by targeting SOCS5. Eur Rev Med Pharmacol Sci (2019) 23(17):7314–26.
193. Zhang Q, Zheng J, Liu L. The long noncoding RNA PCGEM1 promotes cell proliferation, migration and invasion via targeting the miR-182/FBXW11 axis in cervical cancer. Cancer Cell Int (2019) 19(1):1–15. doi: 10.1186/s12935-019-1030-8
194. Shan D, Shang Y, Hu T. MicroRNA-411 inhibits cervical cancer progression by directly targeting STAT3. Oncol Res (2019) 27(3):349–58. doi: 10.3727/096504018X15247361080118
195. Yang S, Zhang X, Sun Y, Shi J, Jiang D, Wang J, et al. MicroRNA-362-3p inhibits migration and invasion via targeting BCAP31 in cervical cancer. Front Mol Biosci (2020) 7:107. doi: 10.3389/fmolb.2020.00107
196. Li T, Zhou W, Li Y, Gan Y, Peng Y, Xiao Q, et al. Correction: MiR-4524b-5p/WTX/β-catenin axis functions as a regulator of metastasis in cervical cancer. PloS One (2019) 14(12):e0226864. doi: 10.1371/journal.pone.0226864
197. Gong Y, Wan JH, Zou W, Lian GY, Qin JL, Wang QM. MiR-29a inhibits invasion and metastasis of cervical cancer via modulating methylation of tumor suppressor SOCS1. Future Oncol (2019) 15(15):1729–44. doi: 10.2217/fon-2018-0497
198. Xu J, Wang H, Wang H, Chen Q, Zhang L, Song C, et al. The inhibition of miR-126 in cell migration and invasion of cervical cancer through regulating ZEB1. Hereditas (2019) 156:11. doi: 10.1186/s41065-019-0087-7
199. Yin S, Zhang Q, Wang Y, Li S, Hu R. MicroRNA-130a regulated by HPV18 E6 promotes proliferation and invasion of cervical cancer cells by targeting TIMP2. Exp Ther Med (2019) 17(4):2837–46. doi: 10.3892/etm.2019.7226
200. Ye C, Hu Y, Wang J. MicroRNA-377 targets zinc finger e-box-Binding homeobox 2 to inhibit cell proliferation and invasion of cervical cancer. Oncol Res (2019) 27(2):183–92. doi: 10.3727/096504018X15201124340860
201. Li N, Cui T, Guo W, Wang D, Mao L. MiR-155-5p accelerates the metastasis of cervical cancer cell via targeting TP53INP1. Onco Targets Ther (2019) 12:3181–96. doi: 10.2147/OTT.S193097
202. Liu YJ, Zhou HG, Chen LH, Qu DC, Wang CJ, Xia ZY, et al. MiR-32-5p regulates the proliferation and metastasis of cervical cancer cells by targeting HOXB8. Eur Rev Med Pharmacol Sci (2019) 23(1):87–95.
203. Qu D, Yang Y, Huang X. miR-199a-5p promotes proliferation and metastasis and epithelial-mesenchymal transition through targeting PIAS3 in cervical carcinoma. J Cell Biochem (2019) 120(8):13562–72. doi: 10.1002/jcb.28631
204. Wu J, Zhao Y, Li F, Qiao B. MiR-144-3p: a novel tumor suppressor targeting MAPK6 in cervical cancer. J Physiol Biochem (2019) 75(2):143–52. doi: 10.1007/s13105-019-00681-9
205. Kapora E, Feng S, Liu W, Sakhautdinova I, Gao B, Tan W. MicroRNA-505-5p functions as a tumor suppressor by targeting cyclin-dependent kinase 5 in cervical cancer. Biosci Rep (2019) 39(7). doi: 10.1042/BSR20191221
206. Chen X, Cao R, Liu H, Zhang T, Yuan X, Xu S. MicroRNA−15a−5p−targeting oncogene YAP1 inhibits cell viability and induces cell apoptosis in cervical cancer cells. Int J Mol Med (2020) 46(4):1301–10.
207. Xia N, Tan WF, Peng QZ, Cai HN. MiR-374b reduces cell proliferation and cell invasion of cervical cancer through regulating FOXM1. Eur Rev Med Pharmacol Sci (2019) 23(2):513–21.
208. Chuang PC, Lu CW, Tsai CC, Tseng SH, Su WH. MicroRNA-128 confers anti-endothelial adhesion and anti-migration properties to counteract highly metastatic cervical cancer cells' migration in a parallel-plate flow chamber. Int J Mol Sci (2020) 22(1). doi: 10.3390/ijms22010215
209. Hu Y, Wu F, Liu Y, Zhao Q, Tang H. DNMT1 recruited by EZH2-mediated silencing of miR-484 contributes to the malignancy of cervical cancer cells through MMP14 and HNF1A. Clin Epigenet (2019) 11(1):186. doi: 10.1186/s13148-019-0786-y
210. Li H, Wang J, Xu F, Wang L, Sun G, Wang J, et al. By downregulating PBX3, miR-526b suppresses the epithelial-mesenchymal transition process in cervical cancer cells. Future Oncol (2019) 15(14):1577–91. doi: 10.2217/fon-2018-0575
211. Sun Q, Yang Z, Li P, Wang X, Sun L, Wang S, et al. A novel miRNA identified in GRSF1 complex drives the metastasis via the PIK3R3/AKT/NF-κB and TIMP3/MMP9 pathways in cervical cancer cells. Cell Death Dis (2019) 10(9):636.
212. Jin Y, Zhou X, Yao X, Zhang Z, Cui M, Lin Y. MicroRNA-612 inhibits cervical cancer progression by targeting NOB1. J Cell Mol Med (2020) 24(5):3149–56. doi: 10.1111/jcmm.14985
213. Chen X, Wu W, Cao X, Zhao X, Li W, Deng C, et al. lncRNA mortal obligate RNA transcript was downregulated in ovarian carcinoma and inhibits cancer cell proliferation by downregulating miRNA-21. J Cell Biochem (2019) 120(7):11949–54. doi: 10.1002/jcb.28478
214. Shao S, Wang C, Wang S, Zhang H, Zhang Y. Hsa_circ_0075341 is up-regulated and exerts oncogenic properties by sponging miR-149-5p in cervical cancer. Biomedicine Pharmacotherapy (2020) 121:109582. doi: 10.1016/j.biopha.2019.109582
215. Wang Y, Wang L, Wang W, Guo X. Overexpression of circular RNA hsa_circ_0001038 promotes cervical cancer cell progression by acting as a ceRNA for miR-337-3p to regulate cyclin-M3 and metastasis-associated in colon cancer 1 expression. Gene (2020) 733:144273. doi: 10.1016/j.gene.2019.144273
216. Liu Z, Mao L, Wang L, Zhang H, Hu X. miR−218 functions as a tumor suppressor gene in cervical cancer. Mol Med Rep (2020) 21(1):209–19.
217. Lin L, Li N, Hu X, Sun J, He Y. Identification of circ_0085616 as an upregulated and oncogenic circular RNA in cervical cancer via the miR-503-5p-Mediated ATXN7L3 activation. Cancer Biotherapy Radiopharmaceuticals (2020). doi: 10.1089/cbr.2020.3865
218. Zhang T, Xue X, Peng H. Therapeutic delivery of miR-29b enhances radiosensitivity in cervical cancer. Mol Ther (2019) 27(6):1183–94. doi: 10.1016/j.ymthe.2019.03.020
219. Hu D, Sun S, Wang Y. MicroRNA-455-5p exerts inhibitory effect in cervical carcinoma through targeting S1PR1 and blocking mTOR pathway. Arch Gynecology Obstetrics (2020) 301(5):1307–15. doi: 10.1007/s00404-020-05536-z
220. Liu J, Li Y, Chen X, Xu X, Zhao H, Wang S, et al. Upregulation of miR-205 induces CHN1 expression, which is associated with the aggressive behaviour of cervical cancer cells and correlated with lymph node metastasis. BMC Cancer (2020) 20(1):1–13. doi: 10.1186/s12885-020-07478-w
221. Wu X, Liu L, Zhang H. miR−802 inhibits the epithelial−mesenchymal transition, migration and invasion of cervical cancer by regulating BTF3. Mol Med Rep (2020) 22(3):1883–91. doi: 10.3892/mmr.2020.11267
222. Ji X, Guo H, Yin S, Du H. miR-139-5p functions as a tumor suppressor in cervical cancer by targeting TCF4 and inhibiting wnt/β-catenin signaling. OncoTargets Ther (2019) 12:7739. doi: 10.2147/OTT.S215796
223. Ming H, Li B, Zhou L, Goel A, Huang C. Long non-coding RNAs and cancer metastasis: Molecular basis and therapeutic implications. Biochim Biophys Acta Rev Cancer (2021) 1875(2):188519. doi: 10.1016/j.bbcan.2021.188519
224. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol (2019) 20(2):69–84. doi: 10.1038/s41580-018-0080-4
225. Heery R, Finn SP, Cuffe S, Gray SG. Long non-coding RNAs: key regulators of epithelial-mesenchymal transition, tumour drug resistance and cancer stem cells. Cancers (2017) 9(4):38. doi: 10.3390/cancers9040038
226. Jia M, Jiang L, Wang YD, Huang JZ, Yu M, Xue HZ. lincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through notch signaling-induced epithelial-mesenchymal transition. Hepatol Res (2016) 46(11):1137–44. doi: 10.1111/hepr.12659
227. Wu D, Ke Y, Xiao R, Liu J, Li Q, Wang Y. Long non-coding RNA GClnc1 knockdown suppresses progression of epithelial ovarian cancer by recruiting FOXC2 to disrupt the NOTCH1/NF-κB/Snail pathway. Exp Cell Res (2021) 399(1):112422. doi: 10.1016/j.yexcr.2020.112422
228. Tao Y, Han T, Zhang T, Ma C, Sun C. LncRNA CHRF-induced miR-489 loss promotes metastasis of colorectal cancer via TWIST1/EMT signaling pathway. Oncotarget (2017) 8(22):36410. doi: 10.18632/oncotarget.16850
229. Kim J, Siverly AN, Chen D, Wang M, Yuan Y, Wang Y, et al. Ablation of miR-10b suppresses oncogene-induced mammary tumorigenesis and metastasis and reactivates tumor-suppressive pathways. Cancer Res (2016) 76(21):6424–35. doi: 10.1158/0008-5472.CAN-16-1571
230. Yang Y, Wang J. Inhibition of MiR-10b restrains the migration and epithelial-mesenchymal transition of lung cells by targeting LATS2 via TAZ pathway. Med Sci Monitor: Int Med J Exp Clin Res (2020) 26:e920275–1. doi: 10.12659/MSM.920275
231. Tan W-X, Sun G, Shangguan M-Y, Gui Z, Bao Y, Li Y-F, et al. Novel role of lncRNA CHRF in cisplatin resistance of ovarian cancer is mediated by miR-10b induced EMT and STAT3 signaling. Sci Rep (2020) 10(1):1–10. doi: 10.1038/s41598-020-71153-0
232. Sun Y, Hu B, Wang Q, Ye M, Qiu Q, Zhou Y, et al. Long non-coding RNA HOTTIP promotes BCL-2 expression and induces chemoresistance in small cell lung cancer by sponging miR-216a. Cell Death Dis (2018) 9(2):1–17. doi: 10.1038/s41419-017-0113-5
233. Shang A, Wang W, Gu C, Chen C, Zeng B, Yang Y, et al. Long chain non-coding RNA HOTTIP enhances IL-6 expression to promotes immune evasion of ovarian cancer cells by promoting the expression of PD-L1 in neutrophils. (2019).
234. Han S, Jin X, Liu Z, Xing F, Han Y, Yu X, et al. The long noncoding RNA HOTTIP promotes breast cancer cell migration, invasiveness, and epithelial–mesenchymal transition via the wnt–β-catenin signaling pathway. Biochem Cell Biol (2019) 97(5):655–64. doi: 10.1139/bcb-2018-0313
235. Wu H, Wei HY, Chen QQ. Long noncoding RNA HOTTIP promotes the metastatic potential of ovarian cancer through the regulation of the miR-615-3p/SMARCE1 pathway. Kaohsiung J Med Sci (2020) 36(12):973–82. doi: 10.1002/kjm2.12282
236. Li H, Yu B, Li J, Su L, Yan M, Zhang J, et al. Characterization of differentially expressed genes involved in pathways associated with gastric cancer. PloS One (2015) 10(4):e0125013. doi: 10.1371/journal.pone.0125013
237. Chen Y, Lu J, Xia L, Xue D, Yu X, Shen D, et al. Testicular orphan receptor 4 promotes tumor progression and implies poor survival through AKT3 regulation in seminoma. Cancer Sci (2018) 109(2):384–94. doi: 10.1111/cas.13461
238. Cristiano BE, Chan JC, Hannan KM, Lundie NA, Marmy-Conus NJ, Campbell IG, et al. A specific role for AKT3 in the genesis of ovarian cancer through modulation of G(2)-m phase transition. Cancer Res (2006) 66(24):11718–25. doi: 10.1158/0008-5472.CAN-06-1968
239. Duan M, Fang M, Wang C, Wang H, Li M. LncRNA EMX2OS induces proliferation, invasion and sphere formation of ovarian cancer cells via regulating the miR-654-3p/AKT3/PD-L1 axis. Cancer Manage Res (2020) 12:2141. doi: 10.2147/CMAR.S229013
240. He SL, Chen YL, Chen QH, Tian Q, Yi SJ. LncRNA KCNQ1OT1 promotes the metastasis of ovarian cancer by increasing the methylation of EIF2B5 promoter. Mol Med (2022) 28(1):112. doi: 10.1186/s10020-022-00521-5
241. Wang Y, Li L, Zhang X, Zhao X. Long non-coding RNA OIP5-AS1 suppresses microRNA-92a to augment proliferation and metastasis of ovarian cancer cells through upregulating ITGA6. J Ovarian Res (2022) 15(1):25. doi: 10.1186/s13048-021-00937-3
242. Sun M, Chen Y, Liu X, Cui Y. LncRNACASC9 promotes proliferation, metastasis, and cell cycle inovarian carcinoma cells through cyclinG1/TP53/MMP7 signaling. Bioengineered (2021) 12(1):8006–19. doi: 10.1080/21655979.2021.1981795
243. Liang H, Yu T, Han Y, Jiang H, Wang C, You T, et al. And invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer (2018) 17(1):1–13.
244. Lai X, Cheng H. LncRNA colon cancer-associated transcript 1 (CCAT1) promotes proliferation and metastasis of ovarian cancer via miR-1290. Eur Rev Med Pharmacol Sci (2018) 22(2):322–8.
245. Chang L, Guo R, Yuan Z, Shi H, Zhang D. LncRNA HOTAIR regulates CCND1 and CCND2 expression by sponging miR-206 in ovarian cancer. Cell Physiol Biochem (2018) 49(4):1289–303. doi: 10.1159/000493408
246. Zhang Y, Ruan F. LncRNA LEF1-AS1 promotes ovarian cancer development through interacting with miR-1285-3p. Cancer Manage Res (2020) 12:687. doi: 10.2147/CMAR.S227652
247. Ding Y, Fang Q, Li Y, Wang Y. Amplification of lncRNA PVT1 promotes ovarian cancer proliferation by binding to miR-140. Mamm Genome (2019) 30(7):217–25. doi: 10.1007/s00335-019-09808-1
248. Ye W, Ni Z, Yicheng S, Pan H, Huang Y, Xiong Y, et al. Anisomycin inhibits angiogenesis in ovarian cancer by attenuating the molecular sponge effect of the lncRNA−Meg3/miR−421/PDGFRA axis. Int J Oncol (2019) 55(6):1296–312.
249. Wang J, Ding W, Xu Y, Tao E, Mo M, Xu W, et al. Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging (Albany NY) (2020) 12(5):4558. doi: 10.18632/aging.102911
250. Duan H, Li X, Chen Y, Wang Y, Li Z. LncRNA RHPN1-AS1 promoted cell proliferation, invasion and migration in cervical cancer via the modulation of miR-299–3p/FGF2 axis. Life Sci (2019) 239:116856. doi: 10.1016/j.lfs.2019.116856
251. Pan L, Meng Q, Li H, Liang K, Li B. LINC00339 promotes cell proliferation, migration, and invasion of ovarian cancer cells via miR-148a-3p/ROCK1 axes. Biomedicine Pharmacotherapy (2019) 120:109423. doi: 10.1016/j.biopha.2019.109423
252. Liang H, Yu M, Yang R, Zhang L, Zhang L, Zhu D, et al. A PTAL-miR-101-FN1 axis promotes EMT and invasion-metastasis in serous ovarian cancer. Mol Therapy-Oncolytics (2020) 16:53–62. doi: 10.1016/j.omto.2019.12.002
253. Sun Q, Li Q, Xie F. LncRNA-MALAT1 regulates proliferation and apoptosis of ovarian cancer cells by targeting miR-503-5p. OncoTargets Ther (2019) 12:6297. doi: 10.2147/OTT.S214689
254. Zou T, Wang PL, Gao Y, Liang WT. Long noncoding RNA HOTTIP is a significant indicator of ovarian cancer prognosis and enhances cell proliferation and invasion. Cancer Biomarkers (2019) 25(2):133–9. doi: 10.3233/CBM-181727
255. Wang L, He M, Fu L, Jin Y. Role of lncRNAHCP5/microRNA-525–5p/PRC1 crosstalk in the malignant behaviors of ovarian cancer cells. Exp Cell Res (2020) 394(1):112129. doi: 10.1016/j.yexcr.2020.112129
256. Wang A, Jin C, Li H, Qin Q, Li L. LncRNA ADAMTS9-AS2 regulates ovarian cancer progression by targeting miR-182-5p/FOXF2 signaling pathway. Int J Biol Macromolecules (2018) 120:1705–13. doi: 10.1016/j.ijbiomac.2018.09.179
257. Gao H, Li X, Zhan G, Zhu Y, Yu J, Wang J, et al. Long noncoding RNA MAGI1-IT1 promoted invasion and metastasis of epithelial ovarian cancer via the miR-200a/ZEB axis. Cell Cycle (2019) 18(12):1393–406. doi: 10.1080/15384101.2019.1618121
258. Zhou S, Xu A, Song T, Gao F, Sun H, Kong X. lncRNA MIAT regulates cell growth, migration, and invasion through sponging miR-150-5p in ovarian cancer. Cancer Biotherapy Radiopharmaceuticals (2020) 35(9):650–60. doi: 10.1089/cbr.2019.3259
259. Liu W, Yang Y-J, An Q. LINC00963 promotes ovarian cancer proliferation, migration and EMT via the miR-378g/CHI3L1 axis. Cancer Manage Res (2020) 12:463. doi: 10.2147/CMAR.S229083
260. Zhang Y, Li P, Zhu M, Guo Y, Yang J. LINC01308 accelerates the malignant progression of ovarian cancer by binding to miRNA-506. Eur Rev Med Pharmacol Sci (2019) 23:3253–60.
261. Yu H, Xu Y, Zhang D, Liu G. Long noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through regulation of miR-612/HOXA13 pathway. Biochem Biophys Res Commun (2018) 503(3):2095–100. doi: 10.1016/j.bbrc.2018.07.165
262. Wang K, Zhao Y, Wang YM. LncRNA MALAT1 promotes survival of epithelial ovarian cancer cells by downregulating miR-145-5p. Cancer Manag Res (2020) 12:11359–69. doi: 10.2147/CMAR.S267355
263. Liu Y, Wang Y, Fu X, Lu Z. Long non-coding RNA NEAT 1 promoted ovarian cancer cells’ metastasis through regulation of miR-382-3p/ROCK 1 axial. Cancer Sci (2018) 109(7):2188–98. doi: 10.1111/cas.13647
264. Mu Y, Li N, Cui Y-L. The lncRNA CCAT1 upregulates TGFβR1 via sponging miR-490-3p to promote TGFβ1-induced EMT of ovarian cancer cells. Cancer Cell Int (2018) 18(1):1–12.
265. Liu M, Shen C, Wang C. Long noncoding RNA LINC01133 confers tumor-suppressive functions in ovarian cancer by regulating leucine-rich repeat kinase 2 as an miR-205 sponge. Am J Pathol (2019) 189(11):2323–39. doi: 10.1016/j.ajpath.2019.07.020
266. Xue F, Xu YH, Shen CC, Qin ZL, Zhou HB. Non-coding RNA LOXL1-AS1 exhibits oncogenic activity in ovarian cancer via regulation of miR-18b-5p/VMA21 axis. Biomedicine Pharmacotherapy (2020) 125:109568. doi: 10.1016/j.biopha.2019.109568
267. Hu X, Li Y, Kong D, Hu L, Liu D, Wu J. Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer. J Cell Physiol (2019) 234(7):10800–8. doi: 10.1002/jcp.27903
268. Zheng M, Hu Y, Gou R, Nie X, Li X, Liu J, et al. Identification three LncRNA prognostic signature of ovarian cancer based on genome-wide copy number variation. Biomedicine Pharmacotherapy (2020) 124:109810. doi: 10.1016/j.biopha.2019.109810
269. Guo L, Chen J, Liu D, Liu L. OIP5-AS1/miR-137/ZNF217 axis promotes malignant behaviors in epithelial ovarian cancer. Cancer Manage Res (2020) 12:6707. doi: 10.2147/CMAR.S237726
270. Wang Y, Wang X, Han L, Hu D. LncRNA MALAT1 regulates the progression and cisplatin resistance of ovarian cancer cells via modulating miR-1271-5p/E2F5 axis. Cancer Manage Res (2020) 12:9999. doi: 10.2147/CMAR.S261979
271. Li H, Zhou Y, Cheng H, Tian J, Yang S. Roles of a TMPO-AS1/microRNA-200c/TMEFF2 ceRNA network in the malignant behaviors and 5-FU resistance of ovarian cancer cells. Exp Mol Pathol (2020) 115:104481. doi: 10.1016/j.yexmp.2020.104481
272. Xu J, Zhang P, Sun H, Liu Y. LINC01094/miR-577 axis regulates the progression of ovarian cancer. J Ovarian Res (2020) 13(1):1–9. doi: 10.1186/s13048-020-00721-9
273. Cao Y, Shi H, Ren F, Jia Y, Zhang R. Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in epithelial ovarian cancer. Exp Cell Res (2017) 359(1):185–94. doi: 10.1016/j.yexcr.2017.07.030
274. Liu X, Li Y, Wen J, Qi T, Wang Y. Long non-coding RNA TTN-AS1 promotes tumorigenesis of ovarian cancer through modulating the miR-139-5p/ROCK2 axis. Biomedicine Pharmacotherapy (2020) 125:109882. doi: 10.1016/j.biopha.2020.109882
275. Sun Z, Gao S, Xuan L, Liu X. Long non-coding RNA FEZF1-AS1 induced progression of ovarian cancer via regulating miR-130a-5p/SOX4 axis. J Cell Mol Med (2020) 24(7):4275–85. doi: 10.1111/jcmm.15088
276. Xu H, Mao H-L, Zhao X-R, Li Y, Liu P-S. MiR-29c-3p, a target miRNA of LINC01296, accelerates tumor malignancy: Therapeutic potential of a LINC01296/miR-29c-3p axis in ovarian cancer. J Ovarian Res (2020) 13(1):1–9. doi: 10.1186/s13048-020-00631-w
277. Duan W, Nian L, Qiao J, Liu N. LncRNA TUG1 aggravates the progression of cervical cancer by binding PUM2. Eur Rev Med Pharmacol Sci (2019) 23(19):8211–8.
278. Yang X, Xin N, Qu HJ, Wei L, Han Z. Long noncoding RNA TUG1 facilitates cell ovarian cancer progression through targeting MiR-29b-3p/MDM2 axis. Anatomical Rec (2020) 303(12):3024–34. doi: 10.1002/ar.24367
279. Luo M, Zhang L, Yang H, Luo K, Qing C. Long non−coding RNA NEAT1 promotes ovarian cancer cell invasion and migration by interacting with miR−1321 and regulating tight junction protein 3 expression. Mol Med Rep (2020) 22(4):3429–39.
280. Chao H, Zhang M, Hou H, Zhang Z, Li N. HOTAIRM1 suppresses cell proliferation and invasion in ovarian cancer through facilitating ARHGAP24 expression by sponging miR-106a-5p. Life Sci (2020) 243:117296. doi: 10.1016/j.lfs.2020.117296
281. Shu C, Yan D, Mo Y, Gu J, Shah N, He J. Long noncoding RNA lncARSR promotes epithelial ovarian cancer cell proliferation and invasion by association with HuR and miR-200 family. Am J Cancer Res (2018) 8(6):981.
282. Xi J, Feng J, Zeng S. Long noncoding RNA lncBRM facilitates the proliferation, migration and invasion of ovarian cancer cells via upregulation of Sox4. Am J Cancer Res (2017) 7(11):2180.
283. Filippov-Levy N, Reich R, Davidson B. The biological and clinical role of the long non-coding RNA LOC642852 in ovarian carcinoma. Int J Mol Sci (2020) 21(15):5237. doi: 10.3390/ijms21155237
284. Wu Y, Deng Y, Guo Q, Zhu J, Cao L, Guo X, et al. Long non-coding RNA SNHG6 promotes cell proliferation and migration through sponging miR-4465 in ovarian clear cell carcinoma. J Cell Mol Med (2019) 23(8):5025–36. doi: 10.1111/jcmm.14359
285. Zhou C, Steplowski TA, Dickens HK, Malloy KM, Gehrig PA, Boggess JF, et al. Estrogen induction of telomerase activity through regulation of the mitogen-activated protein kinase (MAPK) dependent pathway in human endometrial cancer cells. PloS One (2013) 8(2):e55730. doi: 10.1371/journal.pone.0055730
286. Wang D, Wang D, Wang N, Long Z, Ren X. Long non-coding RNA BANCR promotes endometrial cancer cell proliferation and invasion by regulating MMP2 and MMP1 via ERK/MAPK signaling pathway. Cell Physiol Biochem (2016) 40(3-4):644–56. doi: 10.1159/000452577
287. Zhang XJ, Qi GT, Zhang XM, Wang L, Li FF. lncRNA RHPN1-AS1 promotes the progression of endometrial cancer through the activation of ERK/MAPK pathway. J Obstetrics Gynaecology Res (2021) 47(2):533–43. doi: 10.1111/jog.14548
288. Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell (1999) 97(1):17–27. doi: 10.1016/S0092-8674(00)80711-4
289. Kim HJ, Kim LK, Lee SH, Park SA, Eoh KJ, Kim YT. Expression levels of the long noncoding RNA steroid receptor activator promote cell proliferation and invasion and predict patient prognosis in human cervical cancer. Oncol Lett (2018) 16(4):5410–8. doi: 10.3892/ol.2018.9265
290. Xu K, Liu P, Wei W. mTOR signaling in tumorigenesis. Biochim Biophys Acta (BBA)-Reviews Cancer (2014) 1846(2):638–54. doi: 10.1016/j.bbcan.2014.10.007
291. Castellvi J, Garcia A, Ruiz-Marcellan C, Hernández-Losa J, Peg V, Salcedo M, et al. Cell signaling in endometrial carcinoma: phosphorylated 4E-binding protein-1 expression in endometrial cancer correlates with aggressive tumors and prognosis. Hum Pathol (2009) 40(10):1418–26. doi: 10.1016/j.humpath.2008.12.019
292. Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res (2008) 68(3):631–4. doi: 10.1158/0008-5472.CAN-07-5635
293. Park S-A, Kim LK, Kim YT, Heo T-H, Kim HJ. Long non-coding RNA steroid receptor activator promotes the progression of endometrial cancer via wnt/β-catenin signaling pathway. Int J Biol Sci (2020) 16(1):99. doi: 10.7150/ijbs.35643
294. Yuan J-H, Yang F, Wang F, Ma J-Z, Guo Y-J, Tao Q-F, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell (2014) 25(5):666–81. doi: 10.1016/j.ccr.2014.03.010
295. Han F, Wang C, Wang Y, Zhang L. Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s. Am J Cancer Res (2017) 7(4):770.
296. Zheng X, Liu M, Song Y, Feng C. Long noncoding RNA-ATB impairs the function of tumor suppressor miR-126-mediated signals in endometrial cancer for tumor growth and metastasis. Cancer Biotherapy Radiopharmaceuticals (2019) 34(1):47–55. doi: 10.1089/cbr.2018.2565
297. Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood (2009) 113(11):2526–34. doi: 10.1182/blood-2008-06-162164
298. Díaz-Beyá M, Brunet S, Nomdedéu J, Pratcorona M, Cordeiro A, Gallardo D, et al. Located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget (2015) 6(31):31613–27. doi: 10.18632/oncotarget.5148
299. Luo Y, He Y, Ye X, Song J, Wang Q, Li Y, et al. High expression of long noncoding RNA HOTAIRM1 is associated with the proliferation and migration in pancreatic ductal adenocarcinoma. Pathol Oncol Res (2019) 25(4):1567–77. doi: 10.1007/s12253-018-00570-4
300. Rosikiewicz W, Makałowska I. Biological functions of natural antisense transcripts. Acta Biochim Pol (2016) 63(4):665–73.
301. Li Q, Dong C, Cui J, Wang Y, Hong X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res (2018) 37(1):265. doi: 10.1186/s13046-018-0941-x
302. Yuan C, Zhu X, Han Y, Song C, Liu C, Lu S, et al. Elevated HOXA1 expression correlates with accelerated tumor cell proliferation and poor prognosis in gastric cancer partly via cyclin D1. J Exp Clin Cancer Res (2016) 35:15. doi: 10.1186/s13046-016-0294-2
303. Li X, Pang L, Yang Z, Liu J, Li W, Wang D. LncRNA HOTAIRM1/HOXA1 axis promotes cell proliferation, migration and invasion in endometrial cancer. OncoTargets Ther (2019) 12:10997. doi: 10.2147/OTT.S222334
304. Tian C, Su J, Ma Z, Wu Y, Ma H. lncRNA NBAT1 inhibits cell metastasis and promotes apoptosis in endometrial cancer by sponging miR-21-5p to regulate PTEN. Comput Math Methods Med (2022) 2022:9304392. doi: 10.1155/2022/9304392
305. Lai T, Qiu H, Si L, Zhen Y, Chu D, Guo R. Long noncoding RNA BMPR1B-AS1 facilitates endometrial cancer cell proliferation and metastasis by sponging miR-7-2-3p to modulate the DCLK1/Akt/NF-κB pathway. Cell Cycle (2022) 21(15):1599–618. doi: 10.1080/15384101.2022.2060003
306. Jiang Y, Qiao Z, Jiang J, Zhang J. LINC00958 promotes endometrial cancer cell proliferation and metastasis by regulating the miR-145-3p/TCF4 axis. J Gene Med (2021) 23(7):e3345. doi: 10.1002/jgm.3345
307. Dong P, Xiong Y, Yue J, Xu D, Ihira K, Konno Y, et al. Long noncoding RNA NEAT1 drives aggressive endometrial cancer progression via miR-361-regulated networks involving STAT3 and tumor microenvironment-related genes. J Exp Clin Cancer Res (2019) 38(1):1–15. doi: 10.1186/s13046-019-1306-9
308. Zhu H, Jin Y-M, Lyu X-M, Fan L-M, Wu F. Long noncoding RNA H19 regulates HIF-1α/AXL signaling through inhibiting miR-20b-5p in endometrial cancer. Cell Cycle (2019) 18(19):2454–64. doi: 10.1080/15384101.2019.1648958
309. Fang Q, Sang L, Du S. Long noncoding RNA LINC00261 regulates endometrial carcinoma progression by modulating miRNA/FOXO1 expression. Cell Biochem Funct (2018) 36(6):323–30. doi: 10.1002/cbf.3352
310. Wu X, Cai D, Zhang F, Li M, Wan Q. Long noncoding RNA TUSC7 inhibits cell proliferation, migration and invasion by regulating SOCS4 (SOCS5) expression through targeting miR-616 in endometrial carcinoma. Life Sci (2019) 231:116549. doi: 10.1016/j.lfs.2019.116549
311. Zhang K, Cai Y, Zhou Q, Sun H, Wei J. Long non-coding RNA SNHG14 impedes viability, migration and invasion of endometrial carcinoma cells through modulating miR-93-5p/ZBTB7A axis. Cancer Manage Res (2020) 12:9515. doi: 10.2147/CMAR.S257419
312. Xie P, Cao H, Li Y, Wang J, Cui Z. Knockdown of lncRNA CCAT2 inhibits endometrial cancer cells growth and metastasis via sponging miR-216b. Cancer Biomarkers (2018) 21(1):123–33.
313. Wang L, Zhao S, Mingxin Y. LncRNA NR2F1-AS1 is involved in the progression of endometrial cancer by sponging miR-363 to target SOX4. Die Pharmazie-An Int J Pharm Sci (2019) 74(5):295–300.
314. Yang Y, Wu J, Zhou H, Liu W, Wang J, Zhang Q. STAT1-induced upregulation of lncRNA LINC01123 predicts poor prognosis and promotes the progression of endometrial cancer through miR-516b/KIF4A. Cell Cycle (2020) 19(12):1502–16. doi: 10.1080/15384101.2020.1757936
315. Shen CJ, Cheng YM, Wang CL. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J Drug Target (2017) 25(7):637–44. doi: 10.1080/1061186X.2017.1307379
316. Liu H, Zhang L, Ding X, Sui X. LINC00861 inhibits the progression of cervical cancer cells by functioning as a ceRNA for miR−513b−5p and regulating the PTEN/AKT/mTOR signaling pathway. Mol Med Rep (2021) 23(1):1–1.
317. Guo HM, Yang SH, Zhao SZ, Li L, Yan MT, Fan MC. LncRNA NEAT1 regulates cervical carcinoma proliferation and invasion by targeting AKT/PI3K. Eur Rev Med Pharmacol Sci (2018) 22(13):4090–7.
318. Wang X, Wu Q, Xu B, Wang P, Fan W, Cai Y, et al. MiR-124 exerts tumor suppressive functions on the cell proliferation, motility and angiogenesis of bladder cancer by fine-tuning UHRF1. FEBS J (2015) 282(22):4376–88. doi: 10.1111/febs.13502
319. Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta (2016) 1859(1):169–76. doi: 10.1016/j.bbagrm.2015.06.015
320. Shen X, Zhao W, Zhang Y, Liang B. Long non-coding RNA-NEAT1 promotes cell migration and invasion via regulating miR-124/NF-κB pathway in cervical cancer. OncoTargets Ther (2020) 13:3265. doi: 10.2147/OTT.S220306
321. Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell (1997) 91(2):243–52. doi: 10.1016/S0092-8674(00)80406-7
322. Huang W, Cui X, Chen J, Feng Y, Song E, Li J, et al. Long non-coding RNA NKILA inhibits migration and invasion of tongue squamous cell carcinoma cells via suppressing epithelial-mesenchymal transition. Oncotarget (2016) 7(38):62520. doi: 10.18632/oncotarget.11528
323. Makarov SS. NF-kappaB as a therapeutic target in chronic inflammation: recent advances. Mol Med Today (2000) 6(11):441–8. doi: 10.1016/S1357-4310(00)01814-1
324. Wang F, Jiang X, Wang P. NF−κB interaction long non−coding RNA inhibits migration, invasion and epithelial−mesenchymal transition of cervical cancer cells through inhibiting NF−κB signaling pathways. Exp Ther Med (2020) 20(2):1039–47. doi: 10.3892/etm.2020.8752
325. Wang D, You D, Dong J, Liu T. Knockdown of long non-coding RNA LINC00518 inhibits cervical cancer proliferation and metastasis by modulating JAK/STAT3 signaling. Eur Rev Med Pharmacol Sci (2019) 23(2):496–506.
326. Liu YQ, Liu C, Bai Y, Gao J. LncRNA AATBC indicates development and facilitates cell growth and metastasis of cervical cancer as a sponge of miR-1245b-5p. Kaohsiung J Med Sci (2023) 39(2):115–23. doi: 10.1002/kjm2.12628
327. Luan X, Wu L, Zhang H, Wang Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J Gynecol Oncol (2018) 29(6):e95. doi: 10.3802/jgo.2018.29.e95
328. Feng S, Liu W, Bai X, Pan W, Jia Z, Zhang S, et al. Corrigendum to' LncRNA-CTS promotes metastasis and epithelial-to-mesenchymal transition through regulating miR-505/ZEB2 axis in cervical cancer' [Cancer lett. 465 (2019) 105-117]. Cancer Lett (2020) 493:178.
329. Shang C, Wang W, Liao Y, Chen Y, Liu T, Du Q, et al. LNMICC promotes nodal metastasis of cervical cancer by reprogramming fatty acid metabolism. Cancer Res (2018) 78(4):877–90. doi: 10.1158/0008-5472.CAN-17-2356
330. Yang J, Jiang B, Hai J, Duan S, Dong X, Chen C. Long noncoding RNA opa-interacting protein 5 antisense transcript 1 promotes proliferation and invasion through elevating integrin α6 expression by sponging miR-143-3p in cervical cancer. J Cell Biochem (2019) 120(1):907–16. doi: 10.1002/jcb.27454
331. Wang B, Hang J, Li W, Yuan W. Knockdown of LncRNA DLEU2 inhibits cervical cancer progression via targeting miR-128-3p. Onco Targets Ther (2020) 13:10173–84. doi: 10.2147/OTT.S272292
332. Cui L, Nai M, Zhang K, Li L, Li R. lncRNA WT1-AS inhibits the aggressiveness of cervical cancer cell via regulating p53 expression via sponging miR-330-5p. Cancer Manage Res (2019) 11:651. doi: 10.2147/CMAR.S176525
333. Tong R, Zhang J, Wang C, Li X, Yu T, Wang L. LncRNA PTCSC3 inhibits the proliferation, invasion and migration of cervical cancer cells via sponging miR-574-5p. Clin Exp Pharmacol Physiol (2020) 47(3):439–48. doi: 10.1111/1440-1681.13186
334. Lv R, Zhang QW. The long noncoding RNA FTH1P3 promotes the proliferation and metastasis of cervical cancer through microRNA−145. Oncol Rep (2020) 43(1):31–40.
335. Gao F, Feng J, Yao H, Li Y, Xi J, Yang J. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells Nanomedicine Biotechnol (2019) 47(1):776–82. doi: 10.1080/21691401.2019.1577883
336. Chang Q-Q, Chen C-Y, Chen Z, Chang S. LncRNA PVT1 promotes proliferation and invasion through enhancing Smad3 expression by sponging miR-140-5p in cervical cancer. Radiol Oncol (2019) 53(4):443. doi: 10.2478/raon-2019-0048
337. Zou S, Du X, Lin H, Wang P, Li M. Paclitaxel inhibits the progression of cervical cancer by inhibiting autophagy via lncRNARP11-381N20. 2. Eur Rev Med Pharmacol Sci (2018) 22(10):3010–7.
338. Ma Z, Gu G, Pan W, Chen X. LncRNA PCAT6 accelerates the progression and chemoresistance of cervical cancer through up-regulating ZEB1 by sponging miR-543. OncoTargets Ther (2020) 13:1159. doi: 10.2147/OTT.S232354
339. Hu P, Zhou G, Zhang X, Song G, Zhan L, Cao Y. Long non-coding RNA Linc00483 accelerated tumorigenesis of cervical cancer by regulating miR-508-3p/RGS17 axis. Life Sci (2019) 234:116789. doi: 10.1016/j.lfs.2019.116789
340. Li Q, Feng Y, Chao X, Shi S, Liang M, Qiao Y, et al. HOTAIR contributes to cell proliferation and metastasis of cervical cancer via targetting miR-23b/MAPK1 axis. Bioscience Rep (2018) 38(1). doi: 10.1042/BSR20171563
341. Ji N, Wang Y, Bao G, Yan J, Ji S. LncRNA SNHG14 promotes the progression of cervical cancer by regulating miR-206/YWHAZ. Pathology-Research Pract (2019) 215(4):668–75. doi: 10.1016/j.prp.2018.12.026
342. Ou L, Wang D, Zhang H, Yu Q, Hua F. Decreased expression of miR-138-5p by lncRNA H19 in cervical cancer promotes tumor proliferation. Oncol Res Featuring Preclinical Clin Cancer Ther (2018) 26(3):401–10. doi: 10.3727/096504017X15017209042610
343. Zhang X, Zhao X, Li Y, Zhou Y, Zhang Z. Long noncoding RNA SOX21-AS1 promotes cervical cancer progression by competitively sponging miR-7/VDAC1. J Cell Physiol (2019) 234(10):17494–504. doi: 10.1002/jcp.28371
344. Liao L-M, Zhang F-H, Yao G-J, Ai S-F, Zheng M, Huang L. Role of long noncoding RNA 799 in the metastasis of cervical cancer through upregulation of TBL1XR1 expression. Mol Therapy-Nucleic Acids (2018) 13:580–9. doi: 10.1016/j.omtn.2018.10.007
345. Shi W-J, Liu H, Ge Y-F, Wu D, Tan Y-J, Shen Y-C, et al. LINC00673 exerts oncogenic function in cervical cancer by negatively regulating miR-126-5p expression and activates PTEN/PI3K/AKT signaling pathway. Cytokine (2020) 136:155286. doi: 10.1016/j.cyto.2020.155286
346. Xu Y, Zhou W, Zhang C, Liu X, Lv J, Li X, et al. Long non-coding RNA RP11-552M11. 4 favors tumorigenesis and development of cervical cancer via modulating miR-3941/ATF1 signaling. Int J Biol Macromolecules (2019) 130:24–33. doi: 10.1016/j.ijbiomac.2019.02.083
347. Zhu L, Zhang Q, Li S, Jiang S, Cui J, Dang G. Interference of the long noncoding RNA CDKN2B-AS1 upregulates miR-181a-5p/TGFβI axis to restrain the metastasis and promote apoptosis and senescence of cervical cancer cells. Cancer Med (2019) 8(4):1721–30. doi: 10.1002/cam4.2040
348. Wang L, Zhong Y, Yang B, Zhu Y, Zhu X, Xia Z, et al. LINC00958 facilitates cervical cancer cell proliferation and metastasis by sponging miR-625-5p to upregulate LRRC8E expression. J Cell Biochem (2020) 121(3):2500–9. doi: 10.1002/jcb.29472
349. Chen X, Zhang Z, Ma Y, Su H, Xie P, Ran J. LINC02381 promoted cell viability and migration via targeting miR-133b in cervical cancer cells. Cancer Manage Res (2020) 12:3971. doi: 10.2147/CMAR.S237285
350. Zhang L, Liu S-K, Song L, Yao H-R. SP1-induced up-regulation of lncRNA LUCAT1 promotes proliferation, migration and invasion of cervical cancer by sponging miR-181a. Artif Cells Nanomedicine Biotechnol (2019) 47(1):555–63. doi: 10.1080/21691401.2019.1575840
351. Chen X, Xiong D, Yang H, Ye L, Mei S, Wu J, et al. Long noncoding RNA OPA-interacting protein 5 antisense transcript 1 upregulated SMAD3 expression to contribute to metastasis of cervical cancer by sponging miR-143-3p. J Cell Physiol (2019) 234(4):5264–75. doi: 10.1002/jcp.27336
352. Huang L, Gan X, He L, Wang L, Yu J. Silencing of long non−coding RNA NCK1−AS1 inhibits cell proliferation and migration via inhibition of microRNA−134 in cervical cancer. Exp Ther Med (2019) 18(3):2314–22.
353. Liu Y, Guo R, Qiao Y, Han L, Liu M. LncRNA NNT-AS1 contributes to the cisplatin resistance of cervical cancer through NNT-AS1/miR-186/HMGB1 axis. Cancer Cell Int (2020) 20:1–12. doi: 10.1186/s12935-020-01278-9
354. Xu Y, Zhu H, Ma H, Yuan L, Hu Q, Yang L. LINC01305 inhibits malignant progression of cervical cancer via miR-129-5p/Sox4 axis. Am J Trans Res (2020) 12(11):7581.
355. Dou X, Zhou Q, Wen M, Xu J, Zhu Y, Zhang S, et al. Long noncoding RNA FOXD2-AS1 promotes the malignancy of cervical cancer by sponging MicroRNA-760 and upregulating hepatoma-derived growth factor. Front Pharmacol (2019) 10:1700. doi: 10.3389/fphar.2019.01700
356. Ma WG, Shi SM, Chen L, Lou G, Feng XL. SP1-induced lncRNA FOXD3-AS1 contributes to tumorigenesis of cervical cancer by modulating the miR-296-5p/HMGA1 pathway. J Cell Biochem (2021) 122(2):235–48. doi: 10.1002/jcb.29846
357. Wang A-H, Jin C-H, Cui G-Y, Li H-Y, Wang Y, Yu J-J, et al. MIR210HG promotes cell proliferation and invasion by regulating miR-503-5p/TRAF4 axis in cervical cancer. Aging (Albany NY) (2020) 12(4):3205. doi: 10.18632/aging.102799
358. Wu F, Sui Y, Wang Y, Xu T, Fan L, Zhu H. Long noncoding RNA SNHG7, a molecular sponge for microRNA-485, promotes the aggressive behavior of cervical cancer by regulating PAK4. OncoTargets Ther (2020) 13:685. doi: 10.2147/OTT.S232542
359. Liu J, Wu D, Lin X, Hong Y, Wang X, Zheng C, et al. Long non-coding RNA TUG1 sponges microRNA-381-3p to facilitate cell viability and attenuate apoptosis in cervical cancer by elevating MDM2 expression. Life Sci (2021) 267:118902. doi: 10.1016/j.lfs.2020.118902
360. Guo Q, Zhang Q, Lu L, Xu Y. Long noncoding RNA RUSC1-AS1 promotes tumorigenesis in cervical cancer by acting as a competing endogenous RNA of microRNA-744 and consequently increasing bcl-2 expression. Cell Cycle (2020) 19(10):1222–35. doi: 10.1080/15384101.2020.1749468
361. Peng J, Hou F, Feng J, Xu SX, Meng XY. Long non-coding RNA BCYRN1 promotes the proliferation and metastasis of cervical cancer via targeting microRNA-138 in vitro and in vivo. Oncol Lett (2018) 15(4):5809–18. doi: 10.3892/ol.2018.8015
362. Yang T, Xia S. Study of the biological function of LncRNA LUCAT1 on cervical cancer cells by targeting miR-199b-5p. Bioscience Rep (2020) 40(4). doi: 10.1042/BSR20200422
363. Li YJ, Yang Z, Wang YY, Wang Y. Long noncoding RNA ZNF667-AS1 reduces tumor invasion and metastasis in cervical cancer by counteracting microRNA-93-3p-dependent PEG3 downregulation. Mol Oncol (2019) 13(11):2375–92. doi: 10.1002/1878-0261.12565
364. Dong J, Wang Q, Li L, Xiao-Jin Z. Upregulation of long non-coding RNA small nucleolar RNA host gene 12 contributes to cell growth and invasion in cervical cancer by acting as a sponge for MiR-424-5p. Cell Physiol Biochem (2018) 45(5):2086–94. doi: 10.1159/000488045
365. Qi H, Lu L, Wang L. Long noncoding RNA ST7-AS1 upregulates TRPM7 expression by sponging microRNA-543 to promote cervical cancer progression. OncoTargets Ther (2020) 13:7257. doi: 10.2147/OTT.S253868
366. Chen P, Wang R, Yue Q, Hao M. Long non-coding RNA TTN-AS1 promotes cell growth and metastasis in cervical cancer via miR-573/E2F3. Biochem Biophys Res Commun (2018) 503(4):2956–62. doi: 10.1016/j.bbrc.2018.08.077
367. Zhang D, Zhang Y, Sun X. LINC01133 promotes the progression of cervical cancer via regulating miR-30a-5p/FOXD1. Asia-Pacific J Clin Oncol (2020). doi: 10.1111/ajco.13451
368. Li Y, Wang H, Huang H. Long non-coding RNA MIR205HG function as a ceRNA to accelerate tumor growth and progression via sponging miR-122–5p in cervical cancer. Biochem Biophys Res Commun (2019) 514(1):78–85. doi: 10.1016/j.bbrc.2019.04.102
369. Jiang H, Huang G, Zhao N, Zhang T, Jiang M, He Y, et al. Long non-coding RNA TPT1-AS1 promotes cell growth and metastasis in cervical cancer via acting AS a sponge for miR-324-5p. J Exp Clin Cancer Res (2018) 37(1):169. doi: 10.1186/s13046-018-0846-8
370. Jiang H, Liang M, Jiang Y, Zhang T, Mo K, Su S, et al. The lncRNA TDRG1 promotes cell proliferation, migration and invasion by targeting miR-326 to regulate MAPK1 expression in cervical cancer. Cancer Cell Int (2019) 19:152. doi: 10.1186/s12935-019-0872-4
371. Hong H, Zhu H, Zhao S, Wang K, Zhang N, Tian Y, et al. The novel circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression. Cell Death Dis (2019) 10(12):1–19. doi: 10.1038/s41419-019-2183-z
372. Li S, Han Y, Liang X, Zhao M. LINC01089 inhibits the progression of cervical cancer via inhibiting miR-27a-3p and increasing BTG2. J Gene Med (2021) 23(1):e3280. doi: 10.1002/jgm.3280
373. Fan MJ, Zou YH, He PJ, Zhang S, Sun XM, Li CZ. Long non-coding RNA SPRY4-IT1 promotes epithelial-mesenchymal transition of cervical cancer by regulating the miR-101-3p/ZEB1 axis. Biosci Rep (2019) 39(6). doi: 10.1042/BSR20181339
374. Sun J, Chu H, Ji J, Huo G, Song Q, Zhang X. Long non-coding RNA HOTAIR modulates HLA-g expression by absorbing miR-148a in human cervical cancer. Int J Oncol (2016) 49(3):943–52. doi: 10.3892/ijo.2016.3589
375. Ou R, Lv M, Liu X, Lv J, Zhao J, Zhao Y, et al. HPV16 E6 oncoprotein-induced upregulation of lncRNA GABPB1-AS1 facilitates cervical cancer progression by regulating miR-519e-5p/Notch2 axis. FASEB J (2020) 34(10):13211–23. doi: 10.1096/fj.202000762R
376. Zhang Y, Zhang J, Mao L, Li X. Long noncoding RNA HCG11 inhibited growth and invasion in cervical cancer by sponging miR-942-5p and targeting GFI1. Cancer Med (2020) 9(19):7062–71. doi: 10.1002/cam4.3203
377. Wang L, Zhu H. Long non−coding nuclear paraspeckle assembly transcript 1 acts as prognosis biomarker and increases cell growth and invasion in cervical cancer by sequestering microRNA−101. Mol Med Rep (2018) 17(2):2771–7.
378. Yang Z, Ma J, Han S, Li X, Guo H, Liu D. ZFAS1 exerts an oncogenic role via suppressing miR-647 in an m6A-dependent manner in cervical cancer. OncoTargets Ther (2020) 13:11795. doi: 10.2147/OTT.S274492
379. Li P, Wang J, Zhi L, Cai F. Linc00887 suppresses tumorigenesis of cervical cancer through regulating the miR-454-3p/FRMD6-Hippo axis. Cancer Cell Int (2021) 21(1):1–14. doi: 10.1186/s12935-020-01730-w
380. Huo H, Tian J, Wang R, Li Y, Qu C, Wang N. Long non-coding RNA NORAD upregulate SIP1 expression to promote cell proliferation and invasion in cervical cancer. Biomedicine Pharmacotherapy (2018) 106:1454–60. doi: 10.1016/j.biopha.2018.07.101
381. Luo L, Wang M, Li X, Luo C, Tan S, Yin S, et al. A novel mechanism by which ACTA2-AS1 promotes cervical cancer progression: acting as a ceRNA of miR-143-3p to regulate SMAD3 expression. Cancer Cell Int (2020) 20(1):1–13. doi: 10.1186/s12935-020-01471-w
382. Xi J, Feng J, Zeng S, Huang P. Long noncoding RNA UFC 1 is activated by E2F1 and exerts oncogenic properties by functioning as a ce RNA of FOXP 3. Cancer Med (2018) 7(7):3301–10. doi: 10.1002/cam4.1556
383. Zhang L, Guo C, Ji T, Chen X. SOX2 regulates lncRNA CCAT1/MicroRNA-185-3p/FOXP3 axis to affect the proliferation and self-renewal of cervical cancer stem cells. Nanoscale Res Lett (2021) 16(1):1–12. doi: 10.1186/s11671-020-03449-z
384. Chen H, Chi Y, Chen M, Zhao L. Long intergenic non-coding RNA LINC00885 promotes tumorigenesis of cervical cancer by upregulating MACC1 expression through serving as a competitive endogenous RNA for microRNA-432-5p. Cancer Manage Res (2021) 13:1435. doi: 10.2147/CMAR.S291778
385. Li Y. MIR31HG exhibits oncogenic property and acts as a sponge for miR-361-3p in cervical carcinoma. Biochem Biophys Res Commun (2020) 529(4):890–7. doi: 10.1016/j.bbrc.2020.06.028
386. Bao L, Zhong J, Pang L. Upregulation of circular RNA VPS13C-has-circ-001567 promotes ovarian cancer cell proliferation and invasion. Cancer Biother Radiopharm (2019) 34(2):110–8. doi: 10.1089/cbr.2018.2641
387. Zhu X, Li Y, Xie C, Yin X, Liu Y, Cao Y, et al. miR-145 sensitizes ovarian cancer cells to paclitaxel by targeting Sp1 and Cdk6. Int J Cancer (2014) 135(6):1286–96. doi: 10.1002/ijc.28774
388. Lu H, Zheng G, Gao X, Chen C, Zhou M, Zhang L. Propofol suppresses cell viability, cell cycle progression and motility and induces cell apoptosis of ovarian cancer cells through suppressing MEK/ERK signaling via targeting circVPS13C/miR-145 axis. J Ovarian Res (2021) 14(1):1–11. doi: 10.1186/s13048-021-00775-3
389. Zeng B, Zhang X, Zhao J, Wei Z, Zhu H, Fu M, et al. The role of DNMT1/hsa-miR-124-3p/BCAT1 pathway in regulating growth and invasion of esophageal squamous cell carcinoma. BMC Cancer (2019) 19(1):609. doi: 10.1186/s12885-019-5815-x
390. Yang X, Wang J, Li H, Sun Y, Tong X. Downregulation of hsa_circ_0026123 suppresses ovarian cancer cell metastasis and proliferation through the miR−124−3p/EZH2 signaling pathway. Int J Mol Med (2021) 47(2):668–76.
391. Zhang M, Xia B, Xu Y, Zhang Y, Xu J, Lou G, et al. (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif Cells Nanomedicine Biotechnol (2019) 47(1):1224–33. doi: 10.1080/21691401.2019.1593999
392. Xie J, Wang S, Li G, Zhao X, Jiang F, Liu J, et al. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J Cell Mol Med (2019) 23(5):3597–602. doi: 10.1111/jcmm.14260
393. Zhang H, Fan LJ, Liu J, Zhu JQ, Tan TT, Li M, et al. Safflor yellow a protects vascular endothelial cells from ox-LDL-mediated damage. J Recept Signal Transduct Res (2020) 2020:1–8.
394. Hannah J, Zhou P. Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene (2015) 573(1):33–45. doi: 10.1016/j.gene.2015.08.064
395. Duan P-J, Zhao J-J, Xie L-L. Cul4B promotes the progression of ovarian cancer by upregulating the expression of CDK2 and CyclinD1. J Ovarian Res (2020) 13(1):1–10. doi: 10.1186/s13048-020-00677-w
396. Du Z, Wang L, Xia Y. Circ_0015756 promotes the progression of ovarian cancer by regulating miR-942-5p/CUL4B pathway. Cancer Cell Int (2020) 20(1):1–13. doi: 10.1186/s12935-020-01666-1
397. Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J (2017) 284(14):2170–82. doi: 10.1111/febs.14132
398. Pei C, Wang H, Shi C, Zhang C, Wang M. CircRNA hsa_circ_0013958 may contribute to the development of ovarian cancer by affecting epithelial-mesenchymal transition and apoptotic signaling pathways. J Clin Lab Anal (2020) 34(7):e23292. doi: 10.1002/jcla.23292
399. Luo N, Sulaiman Z, Wang C, Ding J, Chen Y, Liu B, et al. Hsa_circ_0000497 and hsa_circ_0000918 contributed to peritoneal metastasis of ovarian cancer via ascites. J Transl Med (2022) 20(1):201. doi: 10.1186/s12967-022-03404-9
400. Li X, Lin S, Mo Z, Jiang J, Tang H, Wu C, et al. CircRNA_100395 inhibits cell proliferation and metastasis in ovarian cancer via regulating miR-1228/p53/epithelial-mesenchymal transition (EMT) axis. J Cancer (2020) 11(3):599. doi: 10.7150/jca.35041
401. Gan X, Zhu H, Jiang X, Obiegbusi SC, Yong M, Long X, et al. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer (2020) 19(1):1–13. doi: 10.1186/s12943-020-01163-z
402. An Q, Liu T, Wang M-y, Yang Y-J, Zhang Z-D, Lin Z-J, et al. circKRT7-miR-29a-3p-COL1A1 axis promotes ovarian cancer cell progression. OncoTargets Ther (2020) 13:8963. doi: 10.2147/OTT.S259033
403. Chen J, Li X, Yang L, Li M, Zhang Y, Zhang J. CircASH2L promotes ovarian cancer tumorigenesis, angiogenesis, and lymphangiogenesis by regulating the miR-665/VEGFA axis as a competing endogenous RNA. Front Cell Dev Biol (2020) 8:595585. doi: 10.3389/fcell.2020.595585
404. Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Therapy-Nucleic Acids (2018) 13:55–63. doi: 10.1016/j.omtn.2018.08.010
405. Zheng Y, Li Z, Yang S, Wang Y, Luan Z. CircEXOC6B suppresses the proliferation and motility and sensitizes ovarian cancer cells to paclitaxel through miR-376c-3p/FOXO3 axis. Cancer Biotherapy Radiopharmaceuticals (2020).
406. Zhang F, Xu Y, Ye W, Jiang J, Wu C. Circular RNA s-7 promotes ovarian cancer EMT via sponging miR-641 to up-regulate ZEB1 and MDM2. Bioscience Rep (2020) 40(7). doi: 10.1042/BSR20200825
407. Deng G, Zhou X, Chen L, Yao Y, Li J, Zhang Y, et al. High expression of ESRP1 regulated by circ-0005585 promotes cell colonization in ovarian cancer. Cancer Cell Int (2020) 20:1–15. doi: 10.1186/s12935-020-01254-3
408. Li M, Cai J, Han X, Ren Y. Downregulation of circNRIP1 suppresses the paclitaxel resistance of ovarian cancer via regulating the miR-211-5p/HOXC8 axis. Cancer Manage Res (2020) 12:9159. doi: 10.2147/CMAR.S268872
409. Xu H, Gong Z, Shen Y, Fang Y, Zhong S. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics (2018) 10(2):187–97. doi: 10.2217/epi-2017-0109
410. Ye Y, Tang X, Sun Z, Chen S. Upregulated WDR26 serves as a scaffold to coordinate PI3K/ AKT pathway-driven breast cancer cell growth, migration, and invasion. Oncotarget (2016) 7(14):17854–69. doi: 10.18632/oncotarget.7439
411. Yuan J, Yin Z, Tao K, Wang G, Gao J. Function of insulin-like growth factor 1 receptor in cancer resistance to chemotherapy. Oncol Lett (2018) 15(1):41–7.
412. Pavelić J, Radaković B, Pavelić K. Insulin-like growth factor 2 and its receptors (IGF 1R and IGF 2R/mannose 6-phosphate) in endometrial adenocarcinoma. Gynecol Oncol (2007) 105(3):727–35. doi: 10.1016/j.ygyno.2007.02.012
413. Bruchim I, Sarfstein R, Werner H. The IGF hormonal network in endometrial cancer: functions, regulation, and targeting approaches. Front Endocrinol (2014) 5:76. doi: 10.3389/fendo.2014.00076
414. Wang Y, Yin L, Sun X. CircRNA hsa_circ_0002577 accelerates endometrial cancer progression through activating IGF1R/PI3K/Akt pathway. J Exp Clin Cancer Res (2020) 39(1):1–16. doi: 10.1186/s13046-020-01679-8
415. Ye X, Chen X. miR-149-5p inhibits cell proliferation and invasion through targeting GIT1 in medullary thyroid carcinoma. Oncol Lett (2019) 17(1):372–8.
416. Liu Y, Chang Y, Cai Y. Hsa_circ_0061140 promotes endometrial carcinoma progression via regulating miR-149-5p/STAT3. Gene (2020) 745:144625. doi: 10.1016/j.gene.2020.144625
417. Hu Q, Du K, Mao X, Ning S. miR-197 is downregulated in cervical carcinogenesis and suppresses cell proliferation and invasion through targeting forkhead box M1. Oncol Lett (2018) 15(6):10063–9. doi: 10.3892/ol.2018.8565
418. Reynolds AB, Roesel DJ, Kanner SB, Parsons JT. Transformation-specific tyrosine phosphorylation of a novel cellular protein in chicken cells expressing oncogenic variants of the avian cellular src gene. Mol Cell Biol (1989) 9(2):629–38.
419. Shibamoto S, Hayakawa M, Takeuchi K, Hori T, Miyazawa K, Kitamura N, et al. Association of p120, a tyrosine kinase substrate, with e-cadherin/catenin complexes. J Cell Biol (1995) 128(5):949–57. doi: 10.1083/jcb.128.5.949
420. Cao N, Mu L, Yang W, Liu L, Liang L, Zhang H. MicroRNA-298 represses hepatocellular carcinoma progression by inhibiting CTNND1-mediated wnt/β-catenin signaling. BioMed Pharmacother (2018) 106:483–90. doi: 10.1016/j.biopha.2018.06.135
421. Shen Q, He T, Yuan H. Hsa_circ_0002577 promotes endometrial carcinoma progression via regulating miR-197/CTNND1 axis and activating wnt/β-catenin pathway. Cell Cycle (2019) 18(11):1229–40. doi: 10.1080/15384101.2019.1617004
422. Wu Y, Wang F, Shi J, Guo X, Li F. CircSMAD2 accelerates endometrial cancer cell proliferation and metastasis by regulating the miR-1277-5p/MFGE8 axis. J Gynecol Oncol (2023) 34(2):e19. doi: 10.3802/jgo.2023.34.e19
423. Shi R, Zhang W, Zhang J, Yu Z, An L, Zhao R, et al. CircESRP1 enhances metastasis and epithelial-mesenchymal transition in endometrial cancer via the miR-874-3p/CPEB4 axis. J Transl Med (2022) 20(1):139. doi: 10.1186/s12967-022-03334-6
424. Zhong L, Zheng C, Fang H, Xu M, Chen B, Li C. MicroRNA-1270 is associated with poor prognosis and its inhibition yielded anticancer mechanisms in human osteosarcoma. IUBMB Life (2018) 70(7):625–32. doi: 10.1002/iub.1753
425. Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol Therapy-Nucleic Acids (2019) 18:24–33. doi: 10.1016/j.omtn.2019.07.012
426. Fardi M, Alivand M, Baradaran B, Farshdousti Hagh M, Solali S. The crucial role of ZEB2: from development to epithelial-to-mesenchymal transition and cancer complexity. J Cell Physiol (2019) 234(9):14783–99. doi: 10.1002/jcp.28277
427. Epifanova E, Babaev A, Newman AG, Tarabykin V. Role of Zeb2/Sip1 in neuronal development. Brain Res (2019) 1705:24–31. doi: 10.1016/j.brainres.2018.09.034
428. Yalim-Camci I, Balcik-Ercin P, Cetin M, Odabas G, Tokay N, Sayan AE, et al. ETS1 is coexpressed with ZEB2 and mediates ZEB2-induced epithelial-mesenchymal transition in human tumors. Mol Carcinogenesis (2019) 58(6):1068–81. doi: 10.1002/mc.22994
429. Feng S, Liu W, Bai X, Pan W, Jia Z, Zhang S, et al. LncRNA-CTS promotes metastasis and epithelial-to-mesenchymal transition through regulating miR-505/ZEB2 axis in cervical cancer. Cancer Lett (2019) 465:105–17. doi: 10.1016/j.canlet.2019.09.002
430. Wang W, Xu A, Zhao M, Sun J, Gao L. Circ_0001247 functions as a miR-1270 sponge to accelerate cervical cancer progression by up-regulating ZEB2 expression level. Biotechnol Lett (2021) 43(3):745–55. doi: 10.1007/s10529-020-03059-w
431. Liu H, Wu Y, Wang S, Jiang J, Zhang C, Jiang Y, et al. Circ-SMARCA5 suppresses progression of multiple myeloma by targeting miR-767-5p. BMC Cancer (2019) 19(1):1–17. doi: 10.1186/s12885-019-6088-0
432. Lu Q, Fang T. Circular RNA SMARCA5 correlates with favorable clinical tumor features and prognosis, and increases chemotherapy sensitivity in intrahepatic cholangiocarcinoma. J Clin Lab Anal (2020) 34(4):e23138. doi: 10.1002/jcla.23138
433. Tan Y, Zhang T, Lian C. Circular RNA SMARCA5 is overexpressed and promotes cell proliferation, migration as well as invasion while inhibits cell apoptosis in bladder cancer. Trans Cancer Res (2019) 8(5):1663–71. doi: 10.21037/tcr.2019.08.08
434. Tian J, Liang L. Involvement of circular RNA SMARCA5/microRNA-620 axis in the regulation of cervical cancer cell proliferation, invasion and migration. Eur Rev Med Pharmacol Sci (2018) 22(24):8589–98.
435. Yu L, Liu X, Cui K, Di Y, Xin L, Sun X, et al. SND1 acts downstream of TGFβ1 and upstream of Smurf1 to promote breast cancer metastasis. Cancer Res (2015) 75(7):1275–86. doi: 10.1158/0008-5472.CAN-14-2387
436. Tseng CW, Yang JC, Chen CN, Huang HC, Chuang KN, Lin CC, et al. Identification of 14-3-3β in human gastric cancer cells and its potency as a diagnostic and prognostic biomarker. Proteomics (2011) 11(12):2423–39. doi: 10.1002/pmic.201000449
437. Zhang X, Zhang Q, Zhang K, Wang F, Qiao X, Cui J. Circ SMARCA5 inhibited tumor metastasis by interacting with SND1 and downregulating the YWHAB gene in cervical cancer. Cell Transplant (2021) 30:0963689720983786. doi: 10.1177/0963689720983786
438. Li H, Hao X, Wang H, Liu Z, He Y, Pu M, et al. Circular RNA expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cell Physiol Biochem (2016) 40(6):1334–44. doi: 10.1159/000453186
439. Zheng S-R, Zhang H-R, Zhang Z-F, Lai S-Y, Huang L-J, Liu J, et al. Human papillomavirus 16 E7 oncoprotein alters the expression profiles of circular RNAs in caski cells. J Cancer (2018) 9(20):3755. doi: 10.7150/jca.24253
440. Meng L, Jia X, Yu W, Wang C, Chen J, Liu F. Circular RNA UBAP2 contributes to tumor growth and metastasis of cervical cancer via modulating miR-361-3p/SOX4 axis. Cancer Cell Int (2020) 20(1):1–13. doi: 10.1186/s12935-020-01436-z
441. Mao Y, Zhang L, Li Y, Yan M, He L. MiR-218 suppresses cell progression by targeting APC in cervical cancer. Int J Clin Exp Pathol (2017) 10(2):2259–69.
442. Li P, Yang X, Cheng Y, Zhang X, Yang C, Deng X, et al. MicroRNA-218 increases the sensitivity of bladder cancer to cisplatin by targeting Glut1. Cell Physiol Biochem (2017) 41(3):921–32. doi: 10.1159/000460505
443. Eoh KJ, Kim HJ, Lee JY, Nam EJ, Kim S, Kim SW, et al. Upregulation of homeobox gene is correlated with poor survival outcomes in cervical cancer. Oncotarget (2017) 8(48):84396–402. doi: 10.18632/oncotarget.21041
444. Mao Y, Zhang L, Li Y. circEIF4G2 modulates the malignant features of cervical cancer via the miR−218/HOXA1 pathway. Mol Med Rep (2019) 19(5):3714–22.
445. Lu C, Liao Z, Cai M, Zhang G. MicroRNA-320a downregulation mediates human liver cancer cell proliferation through the wnt/β-catenin signaling pathway. Oncol Lett (2017) 13(2):573–8. doi: 10.3892/ol.2016.5479
446. Li QQ, Zhang L, Wan HY, Liu M, Li X, Tang H. CREB1-driven expression of miR-320a promotes mitophagy by down-regulating VDAC1 expression during serum starvation in cervical cancer cells. Oncotarget (2015) 6(33):34924–40. doi: 10.18632/oncotarget.5318
447. Zhang C, Han X, Xu X, Zhou Z, Chen X, Tang Y, et al. FoxM1 drives ADAM17/EGFR activation loop to promote mesenchymal transition in glioblastoma. Cell Death Dis (2018) 9(5):469. doi: 10.1038/s41419-018-0482-4
448. Jiang W, Zhou F, Li N, Li Q, Wang L. FOXM1-LDHA signaling promoted gastric cancer glycolytic phenotype and progression. Int J Clin Exp Pathol (2015) 8(6):6756–63.
449. Zhang J, Niu Y, Huang C. Role of FoxM1 in the progression and epithelial to mesenchymal transition of gastrointestinal cancer. Recent Pat Anticancer Drug Discovery (2017) 12(3):247–59. doi: 10.2174/1574892812666170424144352
450. Shi C, Zhang Z. MicroRNA-320 suppresses cervical cancer cell viability, migration and invasion via directly targeting FOXM1. Oncol Lett (2017) 14(3):3809–16. doi: 10.3892/ol.2017.6647
451. Wang W, Luo H, Chang J, Yang X, Zhang X, Zhang Q, et al. Circular RNA circ0001955 promotes cervical cancer tumorigenesis and metastasis via the miR-188-3p/NCAPG2 axis. J Transl Med (2023) 21(1):356. doi: 10.1186/s12967-023-04194-4
452. Song T, Xu A, Zhang Z, Gao F, Zhao L, Chen X, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J Cell Physiol (2019) 234(8):14296–305. doi: 10.1002/jcp.28128
453. Guo J, Chen M, Ai G, Mao W, Li H, Zhou J. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomedicine Pharmacotherapy (2019) 115:108957. doi: 10.1016/j.biopha.2019.108957
454. Jiao J, Jiao X, Liu Q, Qu W, Ma D, Zhang Y, et al. The regulatory role of circRNA_101308 in cervical cancer and the prediction of its mechanism. Cancer Manage Res (2020) 12:4807. doi: 10.2147/CMAR.S242615
455. Wang H, Wei M, Kang Y, Xing J, Zhao Y. Circular RNA circ_PVT1 induces epithelial-mesenchymal transition to promote metastasis of cervical cancer. Aging (Albany NY) (2020) 12(20):20139. doi: 10.18632/aging.103679
456. Fan S, Zhao S, Gao X, Qin Q, Guo Y, Yuan Z, et al. Circular RNA circGSE1 promotes cervical cancer progression through miR-138-5p/Vimentin. OncoTargets Ther (2020) 13:13371. doi: 10.2147/OTT.S282425
457. Ma H, Tian T, Liu X, Xia M, Chen C, Mai L, et al. Upregulated circ_0005576 facilitates cervical cancer progression via the miR-153/KIF20A axis. Biomedicine Pharmacotherapy (2019) 118:109311. doi: 10.1016/j.biopha.2019.109311
458. Zhang S, Chen Z, Sun J, An N, Xi Q. CircRNA hsa_circRNA_0000069 promotes the proliferation, migration and invasion of cervical cancer through miR-873-5p/TUSC3 axis. Cancer Cell Int (2020) 20(1):1–12. doi: 10.1186/s12935-020-01387-5
459. Meng Q-H, Li Y, Kong C, Gao X-M, Jiang X-J. Circ_0000388 exerts oncogenic function in cervical cancer cells by regulating miR-337-3p/TCF12 axis. Cancer Biotherapy Radiopharmaceuticals (2021) 36(1):58–69. doi: 10.1089/cbr.2019.3159
460. Qian W, Huang T, Feng W. Circular RNA HIPK3 promotes EMT of cervical cancer through sponging miR-338-3p to up-regulate HIF-1α. Cancer Manage Res (2020) 12:177. doi: 10.2147/CMAR.S232235
461. Wang J, Li H, Liang Z. Circ-MYBL2 serves as a sponge for miR-361-3p promoting cervical cancer cells proliferation and invasion. OncoTargets Ther (2019) 12:9957. doi: 10.2147/OTT.S218976
Keywords: gynecological cancer, metastasis, invasion, microRNAs: long non-coding RNAs, circular RNAs
Citation: Rezaee A, Ahmadpour S, Jafari A, Aghili S, Zadeh SST, Rajabi A, Raisi A, Hamblin MR, Mahjoubin-Tehran M and Derakhshan M (2023) MicroRNAs, long non-coding RNAs, and circular RNAs and gynecological cancers: focus on metastasis. Front. Oncol. 13:1215194. doi: 10.3389/fonc.2023.1215194
Received: 15 May 2023; Accepted: 28 August 2023;
Published: 03 October 2023.
Edited by:
César López-Camarillo, Universidad Autónoma de la Ciudad de México, MexicoReviewed by:
Jiali Wang, Fourth Hospital of Hebei Medical University, ChinaZongming Li, First Affiliated Hospital of Zhengzhou University, China
Copyright © 2023 Rezaee, Ahmadpour, Jafari, Aghili, Zadeh, Rajabi, Raisi, Hamblin, Mahjoubin-Tehran and Derakhshan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ameneh Jafari, amenehjafari@gmail.com; Maryam Mahjoubin-Tehran, mmahjoubin@gmail.com; Marzieh Derakhshan, mh_derakhshan@yahoo.com
†These authors have contributed equally to this work
 Aryan Rezaee1†
Aryan Rezaee1† Marzieh Derakhshan
Marzieh Derakhshan