- Key Laboratory of State Forestry Administration on Tropical Forestry Research, Research Institute of Tropical Forestry, Chinese Academy of Forestry, Guangzhou, China
Russula griseocarnosa is a wild, ectomycorrhizal, edible, and medicinal fungus with high economic value in southern China. R. griseocarnosa fruiting bodies cannot be artificially cultivated. To better understand the effects of abiotic and biotic factors on R. griseocarnosa growth, the physicochemical properties of R. griseocarnosa and its associated bacterial communities were investigated in two soil types (mycosphere and bulk soil) from Fujian, Guangdong, and Guangxi Provinces. The results revealed that the diversity, community structure, and functional characteristics of the dominant mycosphere bacteria in all geographical locations were similar. Soil pH and available nitrogen (AN) are the major factors influencing the mycosphere–soil bacterial communities’ structure. The diversity of soil bacteria is decreased in R. griseocarnosa mycosphere when compared with the bulk soil. Burkholderia-Paraburkholderia, Mycobacterium, Roseiarcus, Sorangium, Acidobacterium, and Singulisphaera may also be mycorrhiza helper bacteria (MHB) of R. griseocarnosa. The functional traits related to the two-component system, bacterial secretion system, tyrosine metabolism, biosynthesis of unsaturated fatty acids, and metabolism of cofactors and vitamins were more abundant in R. griseocarnosa mycosphere soil. The mycosphere soil bacteria of R. griseocarnosa play a key role in R. griseocarnosa growth. Application of management strategies, such as N fertilizer and microbial fertilizer containing MHB, may promote the conservation, propagation promotion, and sustainable utilization of R. griseocarnosa.
Introduction
Soil is a dynamic biological matrix and an essential part of the terrestrial ecosystem. Soil microbes can participate in crucial processes such as biogeochemical cycles and play a role in different environmental conditions (Cao et al., 2016). Soil bacteria play an influential role in the nitrogen cycle, such as N fixation (Lladó et al., 2017), which is associated with the richness of ectomycorrhizal fungi (Allison et al., 2007; Mediavilla et al., 2019). Soil bacteria, especially mycorrhiza helper bacteria (MHB), can improve the ability of plant roots to form mycorrhiza (Aspray et al., 2006), promote the growth of fungi on soil or root surface, and trigger the germination of fungi in soil (Frey-Klett et al., 2007, 2011). Bacteria may have a variety of symbiotic functions in mushrooms, including inhibiting pathogens and antagonists, improving spore distribution, provisioning of the growth regulators and vitamins (Riedlinger et al., 2006), and increasing mushroom production (Pent et al., 2017). Bacteria were found in fungal hyphae, mycorrhiza, and fungal fruit bodies (Boer et al., 2005; Pent et al., 2017). These MHB serve as biofertilizers to promote fruiting bodies’ formation and increase their productivity (Young et al., 2013). Ectomycorrhizal fungi release many hyphae that contribute to the absorption of water and nutrients (Martin et al., 2007) and can also be used as carriers to transport bacteria (Boer et al., 2005).
Russula griseocarnosa is a wild, edible, medicinal, and ectomycorrhizal symbiont fungi distributed broadly in southern China (Yu et al., 2020). The fruiting bodies of R. griseocarnosa cannot be artificially cultivated (Chen et al., 2010; Ming et al., 2014). R. griseocarnosa has high economic value; its flesh has high nutritional value (Chen et al., 2010; Ming et al., 2014). R. griseocarnosa has been proved to have beneficial effects on dispelling or preventing heart disease and softening brain veins (Chen et al., 2010) when used as a functional food (Chen et al., 2010). R. griseocarnosa polysaccharides have antioxidant activities (Ming et al., 2014) and inhibit the proliferation of cervical cancer cells (Yuan et al., 2017; Liu et al., 2018). Based on the location and the quality of R. griseocarnosa, the fruiting bodies of R. griseocarnosa can sell for $35–$45/kg, while dried of R. griseocarnosa are sold for $140–$180/kg (Ming et al., 2014), with prices increasing. R. griseocarnosa hyphae aggregate densely with the soil around ectomycorrhizal host trees such as Betula platyphylla, Castanopsis carlesii, Pinus massoniana, and Psychotria asiatica. In the symbiotic relationship between fungi and host trees, the fungus can absorb essential elements, especially phosphorus (Hall et al., 2003), to promote the growth of trees, and the trees can provide carbohydrates to the fungus (Giomaro et al., 2005). The fruit body formation of ectomycorrhizal mushrooms must have a symbiotic relationship with plants under certain conditions, and the process is hard to achieve artificially for most of the edible ectomycorrhizal fungi (Hall et al., 2003; Giomaro et al., 2005), such as R. griseocarnosa. There is evidence that several bacteria are selected in the mycosphere of the ectomycorrhizal Laccaria proxima (Warmink et al., 2009). Pseudomonas and Burkholderia are the main bacterial communities in the fruit bodies and in the soil environment of Russula decolorans (Pent et al., 2017). The Pseudomonas communities are significantly increased in the L. proxima mycospheres compared with the corresponding bulk soil (Warmink et al., 2009). Further evidence reveals that bacteria can trigger (Noble et al., 2009) or inhibit (Munsch et al., 2002; Yun et al., 2013) fruiting bodies’ formation of mushrooms. The composition of bacteria within fruiting bodies can be affected directly or indirectly by soil bacterial communities (Antony-Babu et al., 2013), suggesting that R. griseocarnosa may also have helper bacteria to grow and maintain mycelium in the soil.
Soil physicochemical properties, fungi, and other factors may affect the community structure of soil microbial communities (Garbeva et al., 2004). Singh et al. (2008) showed that plant species affect rhizosphere fungi but not rhizosphere bacteria. Soil microbial community and related environmental parameters drive rhizosphere bacterial community structure more than plant genotypes or species (Bulgarelli et al., 2012; Vandenkoornhuyse et al., 2015). The soil contains a variety of bacterial communities shaped by environmental forces (Rousk et al., 2010). These environmental forces may indirectly affect the structure of the bacterial communities in the mycelium and the fruiting bodies of fungi (Warmink et al., 2009; Pent et al., 2017). The effects of bacteria on ectomycorrhizal fungi can vary according to soil factors such as pH and carbon availability (Brulé et al., 2001; Pent et al., 2017; Oh and Lim, 2018). The bacteria in the surrounding soil are filtered by the conditions created by the fruiting bodies, and some bacteria are still retained in the fruiting bodies (Antony-Babu et al., 2013; Pent et al., 2017). MHB are not plant-specific but selective for fungal species (Pivato et al., 2009). This selectivity has been found in fungi that select the soil bacterial communities based on fungal (Halsey et al., 2016) and specific soil properties, such as pH and soil organic carbon (SOC) content (Pent et al., 2017). The non-random selection may depend on their symbiotic functions or habitat requirements (Pent et al., 2017). This selectivity is more conducive to the development of fungal fruiting bodies. Fruiting body formation of L. proxima can be triggered by Pseudomonas communities (Warmink et al., 2009). Bacterial metabolites, nutrients, or stimuli can have a positive or negative effect on fungal growth or spore germination (Oh and Lim, 2018). Leyval and Berthelin (1991) speculated that bacteria could dissolve soil nutrients and cooperate with ectomycorrhizal fungi to increase the diffusion of host roots.
We aimed to explore the characteristics of soil bacteria related to the growth of R. griseocarnosa by comparing the diversity, community structure, and functional profiles of bacteria in the mycosphere and bulk soil. We used Miseq sequencing to expand the research scope and improve the accuracy by comparing soil types in different geographical locations. Also, PICRUSt was used to predict and compare the functional spectrum of bacteria in the mycosphere soil of R. griseocarnosa. We expect this study will not only help us to understand the interaction between R. griseocarnosa and soil bacteria but also provide a theoretical basis for the conservation and propagation of R. griseocarnosa.
Materials and Methods
Sample Collection
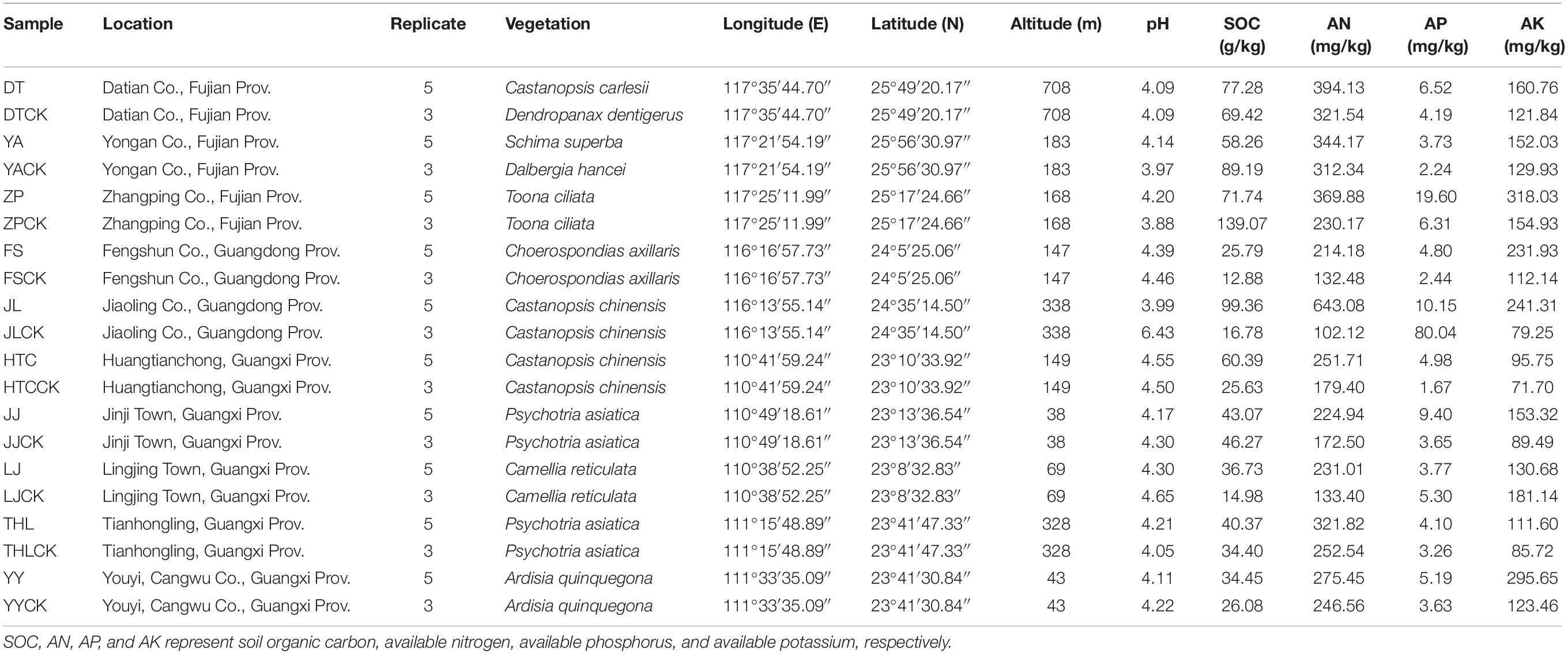
Eighty soil samples from 10 R. griseocarnosa growth sites were collected. Growth sites were distributed in three provinces of China within the longitudinal ranges from 110°38′ to 117°35′ during July 2017 (Table 1). The environment of each site is composed of forest lands with different and distinct vegetation (Table 1). All regions encompass altitude ranges from 38 to 708 m above sea level and a fruiting air temperature range from 21 to 38°C. Geographic distance range from 6.50 to 763.48 km (Supplementary Table S1).
The geographic location and vegetative characteristics are listed in Table 1. At each site, the five R. griseocarnosa fruiting bodies were excavated at a depth of 10 cm using a sterile hand trowel; mycosphere soil was then transferred into a sterile polythene bag (Warmink and van Elsas, 2008; Oh et al., 2016). Samples were collected in the no-fruiting-bodies area with a lateral distance of 40 cm from the R. griseocarnosa and will herein be referred to as “bulk soil” (Warmink and van Elsas, 2008). One fraction of the samples was frozen using liquid nitrogen and stored at −70°C for DNA extraction. The remaining fraction was air-dried and sieved using a 2 mm mesh and then used for physicochemical analysis.
Air-dried samples were used to determine soil pH using a 2 mm mesh with a 1:2.5 (w/v) soil-to-water ratio suspension (Wu et al., 2000). SOC was measured by dichromate oxidation (Nelson and Sommers, 1996). Available phosphorus (AP) was measured using the sodium hydrogen carbonate solution-Mo-Sb anti spectrophotometric (Retamal-Salgado et al., 2017). Soil available potassium (AK) was measured by flame photometry (Zhao et al., 2014). Available nitrogen (AN) was determined by potassium persulfate oxidation (Liu et al., 2015).
DNA Isolation and PCR Amplification
Soil DNA was extracted from 0.30 g soil using the Ezup Column Soil DNA kit (Sangon Biotech, Shanghai) according to the manufacturer instructions (Griffiths et al., 2000). Samples were placed into 1.5 ml centrifuge tubes with 500 mg of glass beads. 400 μl of Buffer SCL at 65°C was added to the samples, followed by incubation at 65°C in a water bath for 5 min. Samples were then centrifuged for 3 min, and the supernatant was collected. An equal volume Buffer SP was added to the supernatant and incubated on ice for 10 min. Following incubation, 200 μl of β-Mercaptoethanol was added, and samples were further centrifuged for 3 min. The supernatant was collected, and 1.5 volumes of Buffer SB were added. Samples were washed twice with 700 and 300 μl Wash Solution, respectively. Finally, 80 μl TE Buffer was added to the center of the adsorption membrane, and the DNA solution was obtained by centrifugation at 12,000 rpm for 3 min. DNA concentration and purity were measured by NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, United States).
The V3-V4 regions of bacterial 16S were amplified by primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Mori et al., 2014). The PCR reactions were conducted using the following program: 95°C for 3 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 45 s, and a final extension of 72°C for 10 min in a GeneAmp 9700 thermocycler PCR system. PCR reactions were performed as follows: 4 μl 5 × FastPfu buffer, 2 μl 2.5 mM dNTPs, 0.8 μl of each primer (5 μM), 0.4 μl FastPfu polymerase, 0.2 μl 2.0 g/l BSA, 2 μl 50 mg/l template DNA, and 9.8 μl ddH2O in a 20 μl total volume. All PCR products were collected from 2% agarose gels and purified using a DNA gel extraction kit (Axygen Biosciences, Inc., United States) and quantified before sequencing.
Miseq Sequencing
Purified products were assembled in an equal volume and sequenced (2 × 300 bp) using Illumina’s Miseq platform in Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China. The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: PRJNA553654).
Bioinformatic Analysis of the 16SrRNA Amplicons
Raw fastq were demultiplexed, quality-filtered, and merged using the following standards: (1) truncate the 300 bp reads where the average quality score <20 over a 50 bp; the truncated read codes less than 50 bp were discarded; (2) precise barcode matching sequences were included, and two nucleotide mismatch in primer matching or reads containing ambiguous characters were deleted; (3) only assemble overlapped sequences exceeding 10 bp according to overlapped sequences; and (4) unassembled readings were discarded.
Operational taxonomic units (OTUs) were clustered at 97% similarity cutoff value, and chimeric sequences were identified and removed using USEARCH1 (version 7.0). The 16S rRNA gene sequence was analyzed by SILVA (SSU123) database using a confidence threshold of 70% (Cole et al., 2013; Quast et al., 2013). The subsampling was based on the minimum sample sequence with equal sequencing depth (16,175 sequences per followed by clustering) (Ye et al., 2017). Diversity metrics, that is, richness (observed species), Chao richness index, Shannon diversity index, and coverage and phylogenetic diversity were calculated based on OTU tables using mother (v.1.30.1). The indexes describe the structure of bacterial communities.
Statistical Analysis
The statistical analysis was conducted using the online platform of Majorbio I-Sanger Cloud Platform2. The results of the two groups of data were consistent with the normal distribution, and the variance of the two groups was not equal. Therefore, the results were expressed as mean values and two-group statistical analyses using Welch’s t-test (Delacre et al., 2017). The bar of diversity index represents the mean ± standard error. Significant correlations are expressed as: ∗ 0.01 < p ≤ 0.05; ∗∗ 0.001 < p ≤ 0.01; ∗∗∗p ≤ 0.001.
LEfSe was used to identify taxa that differed consistently using the default parameters (LDA Score >2, p < 0.05). LEfSe was applied in the identification of mycosphere and bulk soil biomarkers of microbiomes at the genus levels. The biomarkers were classified according to their statistical significance. The results were visualized by using bar charts and cladograms (Segata et al., 2011).
Mantel Tests (1967) with 999 permutations were used to test the Bray–Curtis correlation between soil/site properties and bacterial community structure by QIIME (Caporaso et al., 2010). ANOSIM analysis of the relationship of sites was performed using R’s Vegan package (version 3.3.3) (Oksanen et al., 2017). To analyze the relationship between taxa and the soil/site properties, variation portioning analysis (VPA) was done using R’s Vegan package (Oksanen et al., 2017). The OTUs and soil/site properties were used in the analysis. Detrended correspondence analysis (DCA) was done based on OTUs. Principle component analysis (PCA) plot was drawn by R’s Vegan package (Oksanen et al., 2017).
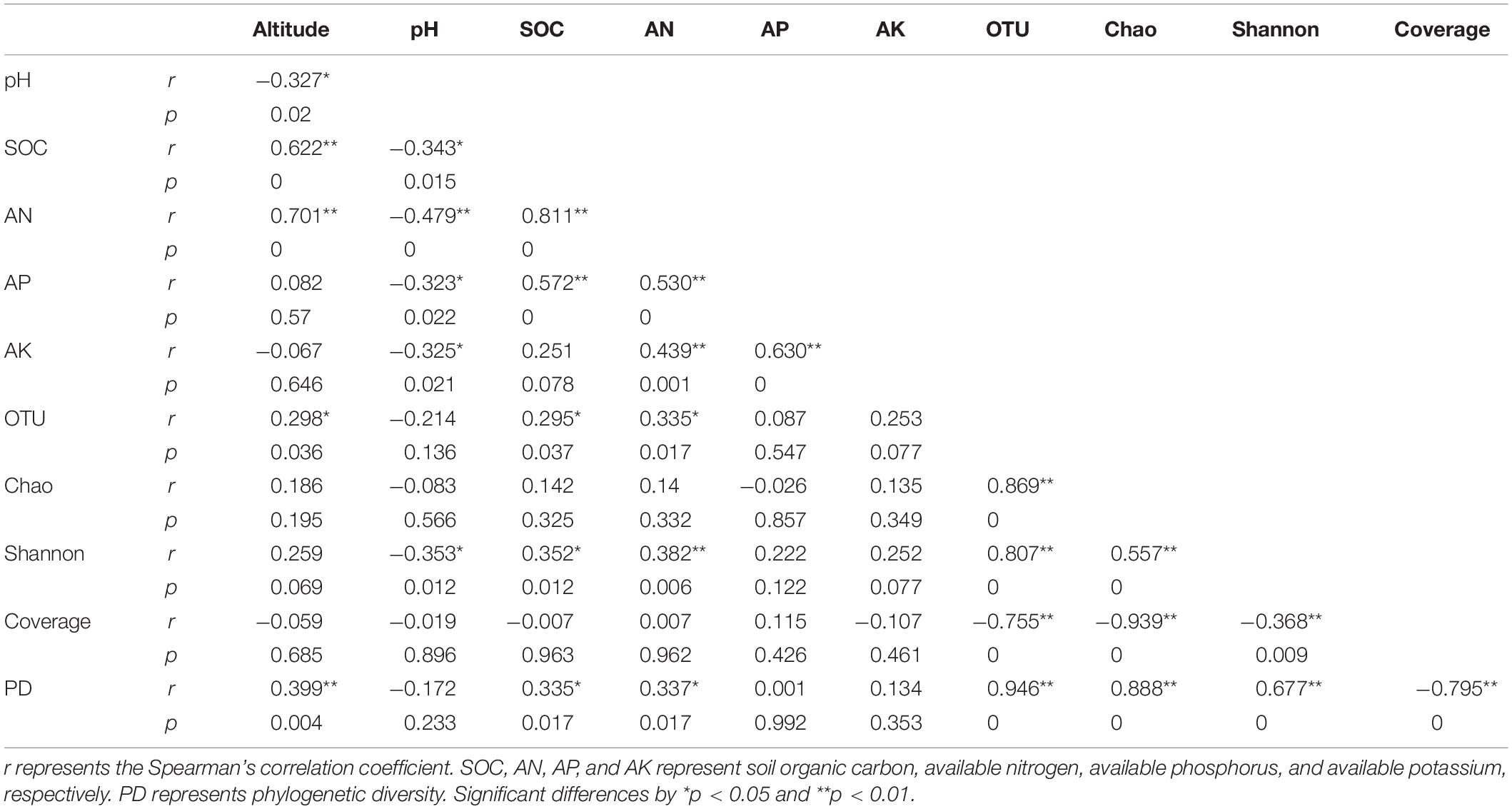
Spearman’s correlation coefficients among the top 30 mycosphere’s bacterial genera and soil properties were calculated and displayed as a heat map using R’s pheatmap package (Kolde, 2019). The Spearman’s correlation analysis of soil properties and the diversity indexes were calculated by SPSS21.0.
16S rRNA Functional Predictions
The microbial function was predicted by PICRUSt (Langille et al., 2013; Oh et al., 2016). OTUs was assigned with QIIME’s command “pick_closed_otus” with 97% similarity in Greengenes13.5 database. Then, the predicted functions were blasted to the KEGG (Kyoto Encyclopedia of Genes and Genomes) database, and statistical differences among groups were compared by STAMP software (Parks and Beiko, 2010). Welch’s t-test and Storey False Discovery Rate (FDR, p < 0.05) were performed for two groups (Storey, 2007).
Results
Site Sampling of Mycosphere and Bulk Soil
Soil organic carbon at the collection sites ranged from 12.88 to 139.07 g/kg (Table 1). Soil pH was between 3.88 and 6.43 at the collection sites. The soil contents of available nitrogen (AN, 102.12–643.08 mg/kg), available phosphorus (AP, 1.67–80.04 mg/kg), and available potassium (AK, 71.70–318.03 mg/kg) showed rich changes in collection sites (Table 1). The geographical distance ranges from 6.50 to 763.48 km (Supplementary Table S1).
Of all sites, the soil pH of YA (p = 0.014) and ZP (p = 0.001) was significantly higher in the mycosphere soil, while soil pH of LJ (p < 0.001), JL (p < 0.001) and JJ (p = 0.032) were significantly lower in the mycosphere soil. The SOC of LJ (p = 0.016), HTC (p = 0.022), and JL (p = 0.010) was significantly higher in the mycosphere soil, while the SOC of YA (p = 0.027) and ZP (p = 0.001) was significantly lower in the mycosphere soil. The AN of LJ (p = 0.041), DT (p = 0.006), JL (p = 0.018), and ZP (p < 0.001) was significantly higher in the mycosphere soil. The AP of YA (p = 0.004) and ZP (p = 0.044) was significantly higher in the mycosphere soil. The AK of JJ (p = 0.020), YA (p = 0.028), and ZP (p < 0.001) was significantly higher in the mycosphere soil. In most sites with mycorrhiza soil, the content of AN, AK, and AP was significantly higher than those of bulk soil. The results showed that mycosphere soils were more nutrient-rich compared with bulk soils (Supplementary Table S2).
Bacteria Communities and Structure in Mycosphere and Bulk Soil
Diversity of Bacterial Community
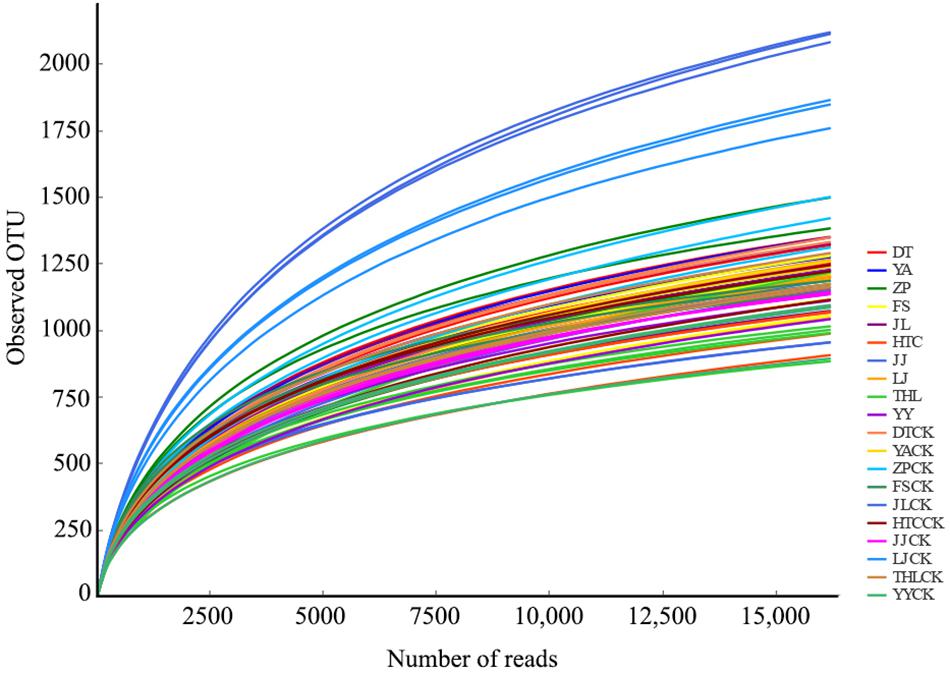
Each sample had 16,175 bacterial sequences for further analysis (Figure 1). A total of 6,014 OTUs were delineated at a 97% similarity level. We investigated the distinctiveness between mycosphere and bulk bacterial communities with samples from ten different sites. Chao and Shannon indexes of mycosphere samples from JL, LJ, and THL were significantly lower than in bulk soil (Figure 2). The Chao index of HTC site (p = 0.010) and ZP site (p = 0.010) was significantly lower than that of bulk soil, while the Shannon index showed no significant difference in bulk soil. Only four sites reported no significant difference between the Chao and Shannon indexes in regard to mycosphere and bulk soil. The bacterial community structure clustered significantly with soil compartments in ten sites (ANOSIM; bacteria: R = 0.74, p = 0.001).
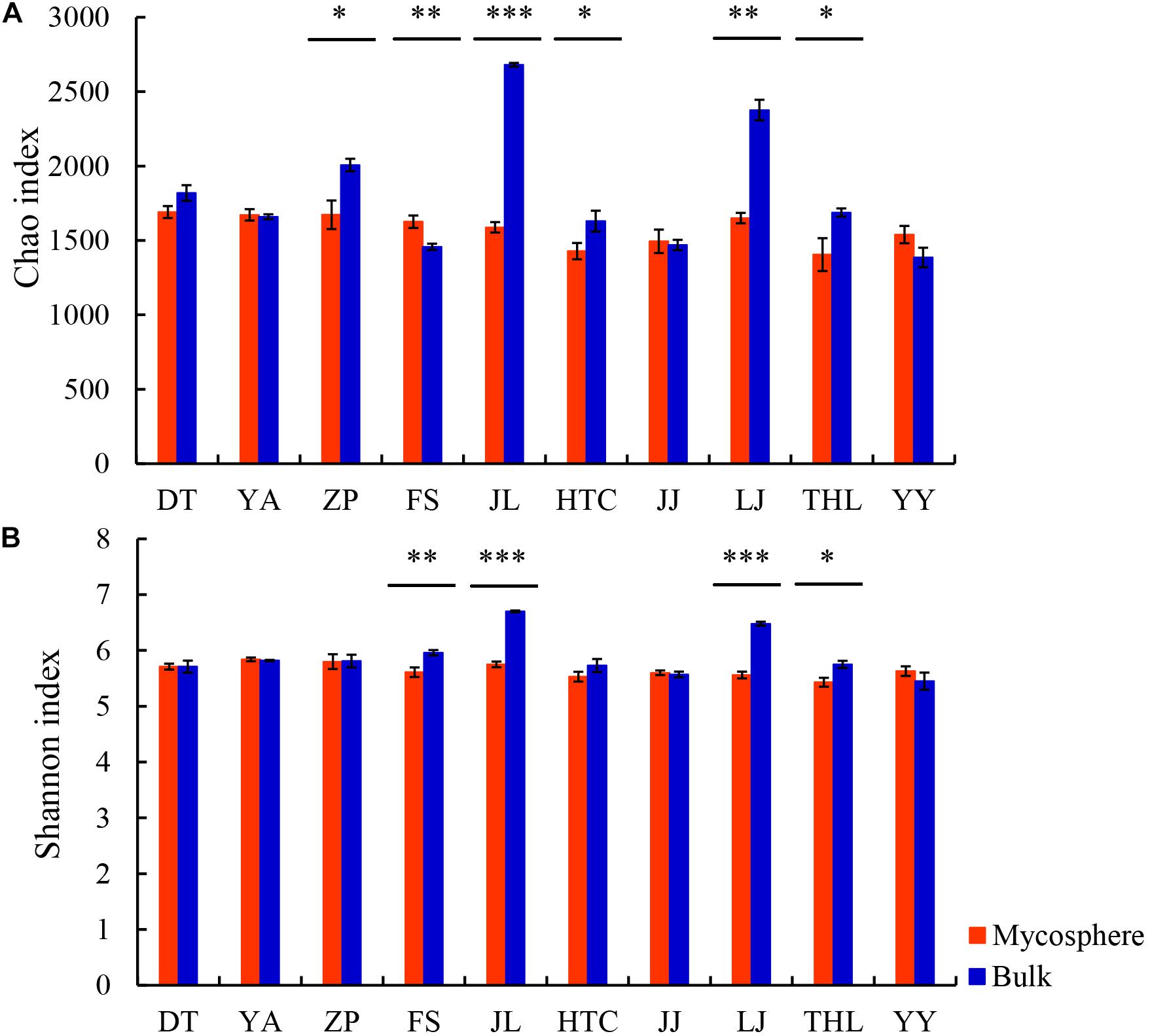
Figure 2. Comparison of Chao (A) and Shannon (B) indexes between mycosphere and bulk soil. Significant differences by *p < 0.05; **p < 0.01 and ***p < 0.001.
Keystone Species in Mycosphere and Bulk Soils
There was a total of 6,014 bacterial OTUs obtained from the ten sites, clustered into 38 phyla. Proteobacteria, Acidobacteria, Actinobacteria, and Chloroflexi were the dominant phyla present in soil samples (Figure 3A), accounting for 86.99 and 86.53% of the total species in mycosphere and bulk soil samples, respectively (Figure 3B). Cyanobacteria, Saccharibacteria, Gemmatimonadetes, and Nitrospirae phyla were also present in all samples examined but at a lower species richness. Proteobacteria (p = 0.023), Planctomycetes (p = 0.012), and Verrucomicrobia (p = 0.034) were significantly higher in mycosphere soil, while Chloroflexi (p = 0.006), Firmicutes (p = 0.040), Cyanobacteria (p = 0.033), Saccharibacteria (p = 0.002), and Gemmatimonadetes (p = 0.006) were significantly lower in mycosphere soil (Figure 3C).
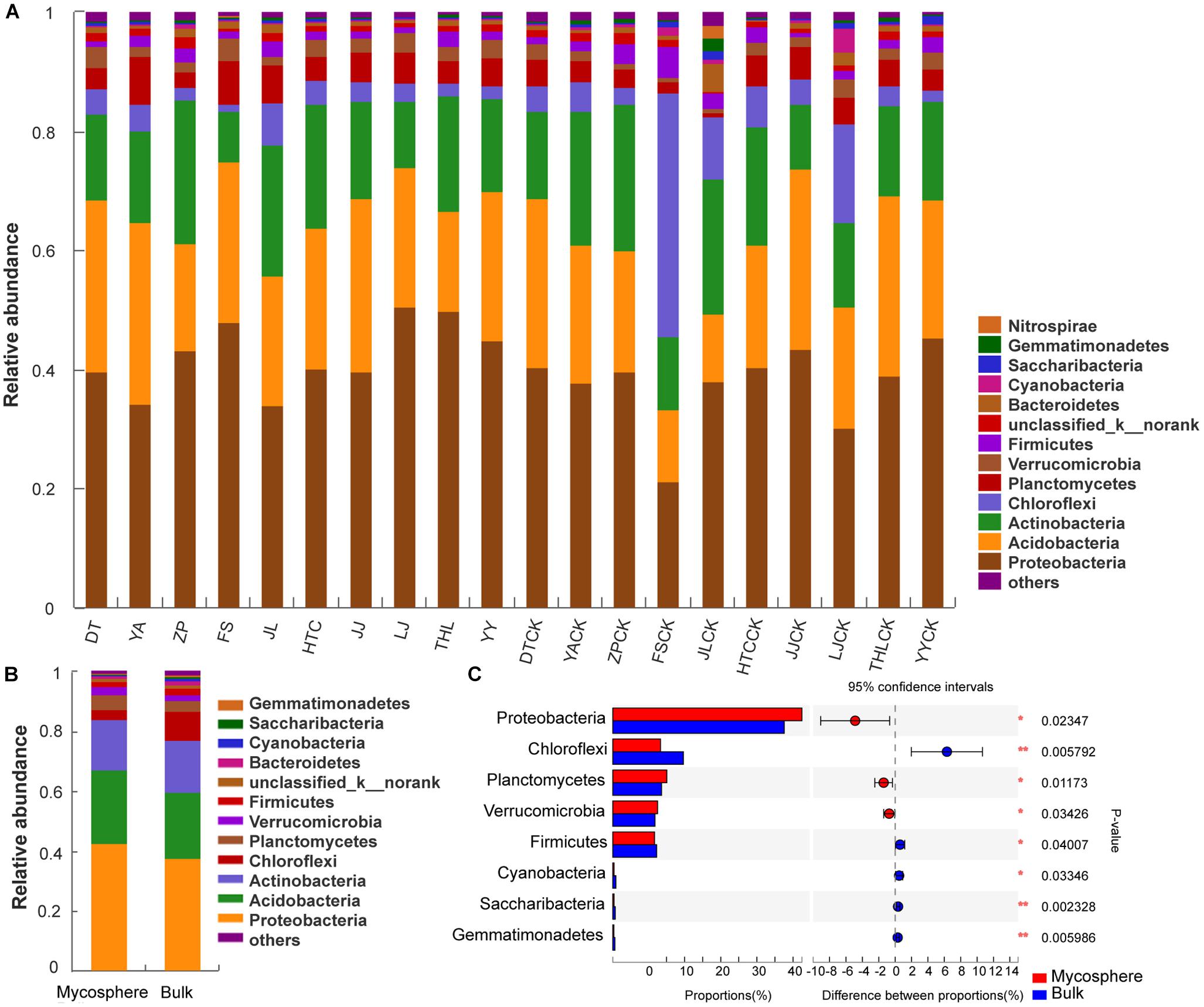
Figure 3. Comparison of phyla between mycosphere and bulk soil. (A) The abundances of phyla of each site. (B) Comparison of the average abundance of phylum in mycosphere and bulk soil. (C) Significant differences among the abundances of phyla between mycosphere and bulk soil. Significant differences by *p < 0.05 and **p < 0.01.
At the phylum level, the relative abundances of Acidobacteria (p = 0.022) and Planctomycetes (p = 0.016) were significantly enriched in YA mycosphere soil, while Actinobacteria (p = 0.015), Saccharibacteria (p = 0.013), and Gemmatimonadetes (p = 0.030) were significantly higher in the YA bulk soil (Supplementary Table S3). The relative abundances of Proteobacteria (p = 0.004), Acidobacteria (p = 0.005), Planctomycetes (p = 0.030), and Verrucomicrobia (p = 0.017) were significantly higher in FS mycosphere soil, while Chloroflexi (p < 0.001), Actinobacteria (p = 0.011), Firmicutes (p = 0.008), and Cyanobacteria (p < 0.001) were significantly higher in FS bulk soil (Supplementary Table S3). At the phylum level, the relative abundances of Acidobacteria (p = 0.015) and Planctomycetes (p = 0.019) were significantly higher in JL mycosphere soil, while Bacteroidetes (p = 0.013), Saccharibacteria (p = 0.022), Gemmatimonadetes (p < 0.001), and Nitrospirae (p = 0.012) were significantly lower (Supplementary Table S3). The relative abundances of Proteobacteria (p = 0.006) were significantly higher in LJ mycosphere soil, while Chloroflexi (p < 0.001), Cyanobacteria (p = 0.017), and Bacteroidetes (p = 0.018) were significantly higher in LJ bulk soil (Supplementary Table S3). The relative abundances of Gemmatimonadetes (p = 0.001) were significantly higher in the HTC bulk soil (Supplementary Table S3). The relative abundances of Acidobacteria (p = 0.046) were significantly higher in the THL bulk soil. These results show that Proteobacteria, Acidobacteria, Planctomycetes, and Verrucomicrobia were significant higher in mycosphere soil, which was consistent with the overall analysis (Supplementary Table S3).
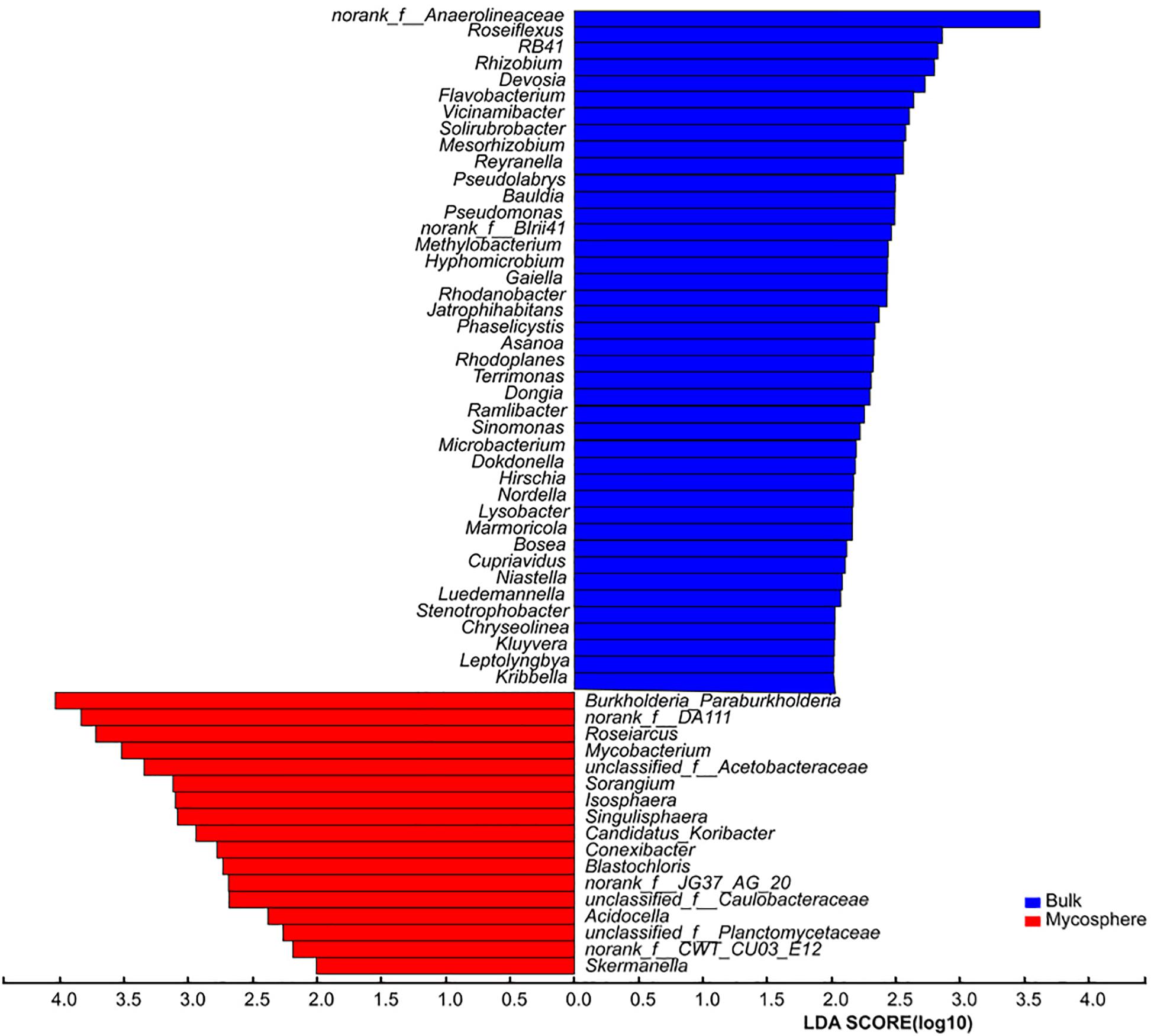
Over 700 genera were found in the sequencing data. The relative abundance of 92 bacterial genera was over 1%. In top 30 genera, the norank_f__DA111 (p = 0.039), Burkholderia-Paraburkholderia (p = 0.045), Mycobacterium (p = 0.025), Roseiarcus (p < 0.001), Candidatus_Xiphinematobacter (p = 0.032), Sorangium (p = 0.019), Acidobacterium (p = 0.020), and Singulisphaera (p = 0.008) were significantly higher in mycosphere soil samples (Figure 4 and Supplementary Table S4), while the norank_c__JG37-AG-4 (p = 0.015) and norank_f__Anaerolineaceae (p = 0.003) were significantly higher in bulk soil (Figure 4). For all genera, mycosphere and bulk soil groups were represented by cladograms, and the LDA scores of two were proved by LEfSe (Figure 5).
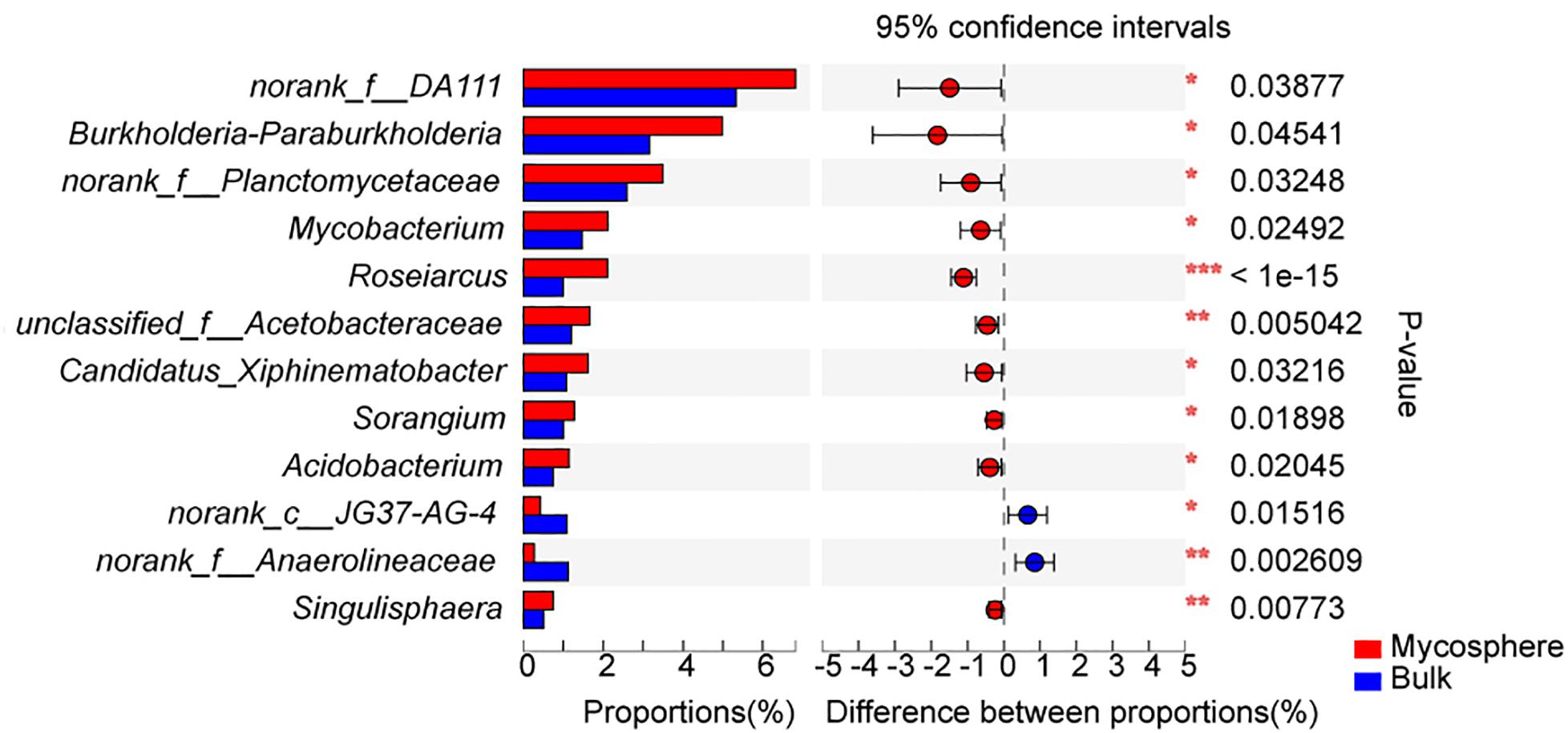
Figure 4. Significant differences among the top 30 genera between mycosphere and bulk soil. Significant differences by *p < 0.05; **p < 0.01 and ***p < 0.001.
Abiotic and Biotic Factors in R. griseocarnosa Mycosphere and Bulk Soils
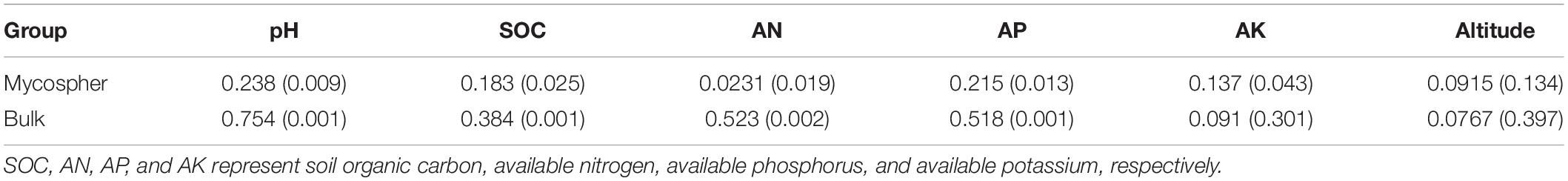
Soil pH, SOC, AN, AP, and AK produce the highest variability in bacterial community structures for both mycosphere and bulk soil, as demonstrated by the Mantel test (Table 2). To quantify the effects of the soil properties and the altitude on mycosphere bacterial communities, a variance partitioning analysis (VPA) was performed. A matrix of the soil properties’ relationship with the soil bacterial community was constructed using RDA analysis.
Correlation analysis showed that there was a significant correlation between the soil parameters and the soil bacterial community structure. These variables explain the changes in bacterial community structure in the mycosphere (24.30%) and bulk soil (39.69%) (Figure 6). Soil parameters constituted 20.56%, altitude constituted 3.71%, and interactions between the soil parameters and altitude explained 0.03% of the variations in the mycosphere bacterial communities (Figure 6A). Meanwhile, for bulk soil, soil parameters explained 33.86%, altitude explained 5.68%, and interactions between the soil parameters and altitude explained the 0.15% of the variations in bacterial communities (Figure 6B). The soil pH and AN were identified as the main contributing factors to the soil parameter and explained the bacterial communities’ variety in the mycosphere at 3.87 and 4.37%, respectively (Figure 6).
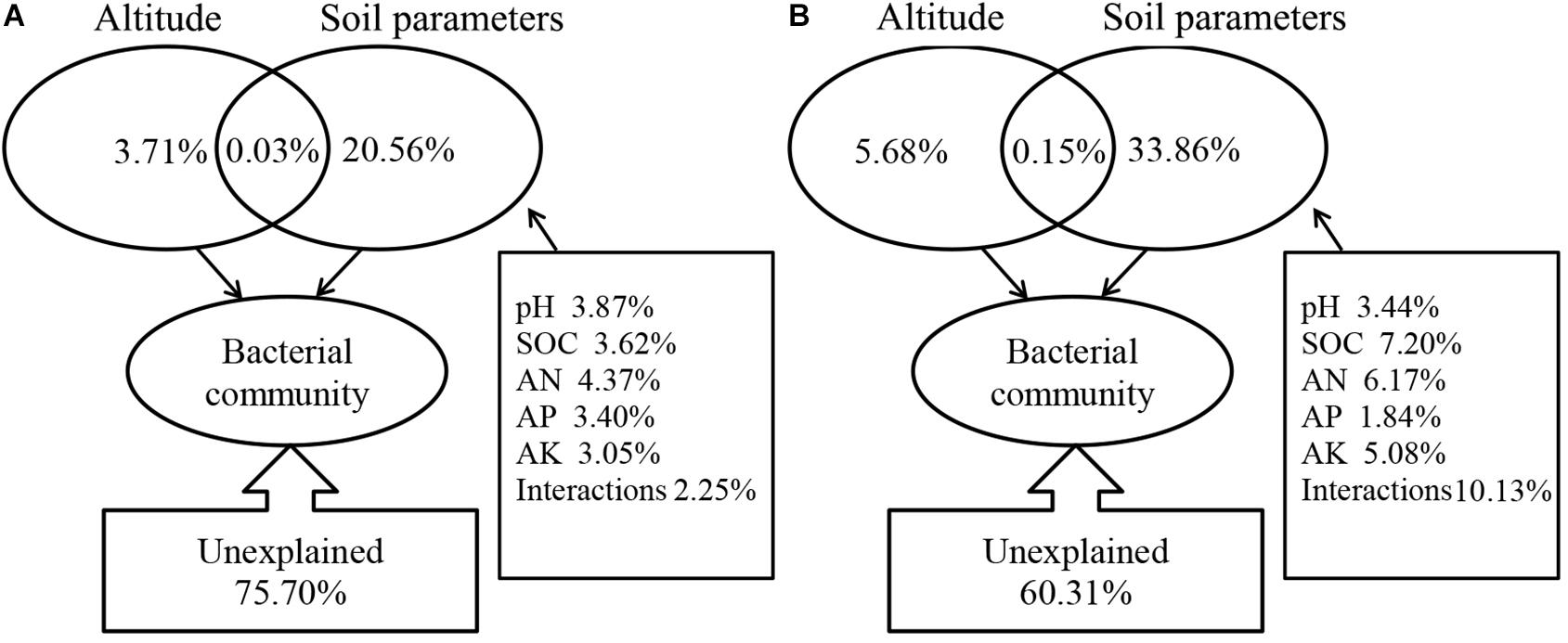
Figure 6. Variation partition analysis (VPA) of soil/site properties on the bacterial community. (A) Mycosphere soil. (B) Bulk soil.
To explore the effect of host plants on soil bacterial, we analyzed the mycosphere bacterial communities of R. griseocarnosa under different host plants by PCA. The first two axes of the PCA explained 20.96 and 13.24% of the variance in the OTU data, respectively. PCA showed that the samples were dispersed among different host plants (Figure 7). It indicates that the host plant had little effect on soil mycosphere bacteria. There were no significant differences in the bacterial diversity index among the five replicates in each square (data not shown), which indicates that the host plant individual has a minimal effect on bacterial diversity.
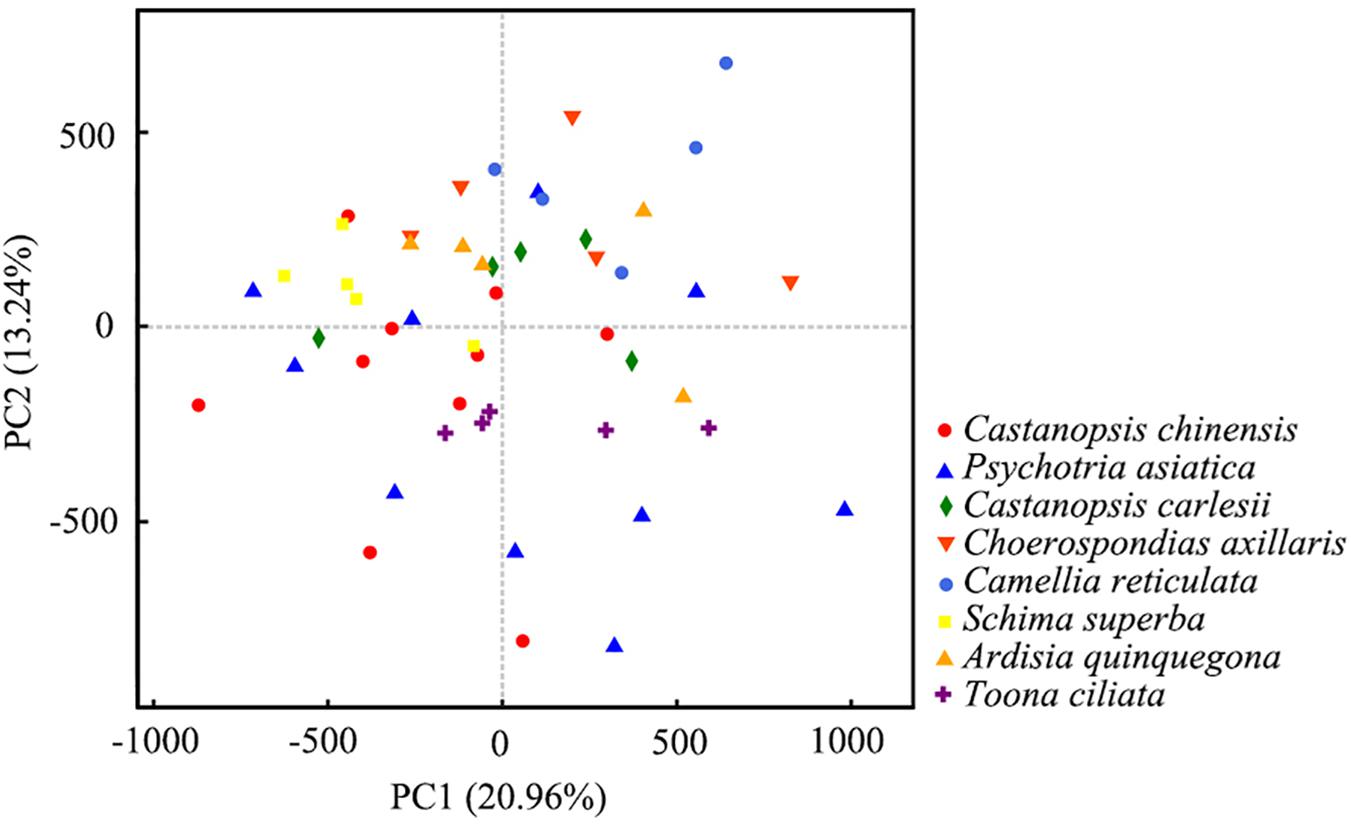
Figure 7. Principle component analysis (PCA) plot of the host plant and soil bacterial communities’ richness. The values of PC1 and PC2, explaining 20.96 and 13.24% of the variance.
Environmental Factors Influence the Mycosphere’s Soil Bacteria Communities
The diversity index was significantly correlated with soil and site properties (Table 3). The OTUs and phylogenetic diversity had a positive correlation with geological location altitude, SOC, and AN (Table 3). The Shannon index was significantly and positively correlated with SOC (p = 0.012) and AN (p = 0.006), while negatively correlated with pH (p = 0.012) (Table 3). Collection mycosphere sites had an acidic soil with sample pH values ranging from 3.99 to 4.55.
The relative abundance of the top 30 genera and soil/site properties was examined by Spearman correlation analysis (Figure 8). The heatmap showed that AP and AK clustered together and altitude, SOC, and AN clustered together, while pH was further apart on the ordination (Figure 8). Variibacter showed a significant positive correlation with pH (p < 0.001) and a significant negative correlation with altitude (p = 0.002), SOC (p = 0.029), and AN (p = 0.003). Acidibacter showed a negative correlation with altitude (p < 0.001) and AN (p = 0.021). Burkholderia-Paraburkholderia showed a significant positive correlation with pH (p = 0.005) and a significant negative correlation with SOC (p = 0.018). Candidatus_Xiphinematobacter presented a negative correlation with AP (p = 0.005), SOC (p = 0.004), and AN (p = 0.021). Acidothermus showed a significant positive correlation with AP (p < 0.001), AK (p = 0.015), SOC (p < 0.001), and AN (p = 0.002) and a significant negative correlation with pH (p = 0.042). Rhizomicrobium showed positive correlation with AP (p < 0.001), AK (p < 0.001), and AN (p = 0.010). Roseiarcus showed a positive correlation with AP (p = 0.001) and AK (p = 0.049). Candidatus_Koribacter showed a significant positive correlation with AP (p = 0.043). Bradyrhizobium showed a significant positive correlation with pH (p = 0.0093). Singulisphaera showed a significant negative correlation with pH (p = 0.017) (Figure 8).
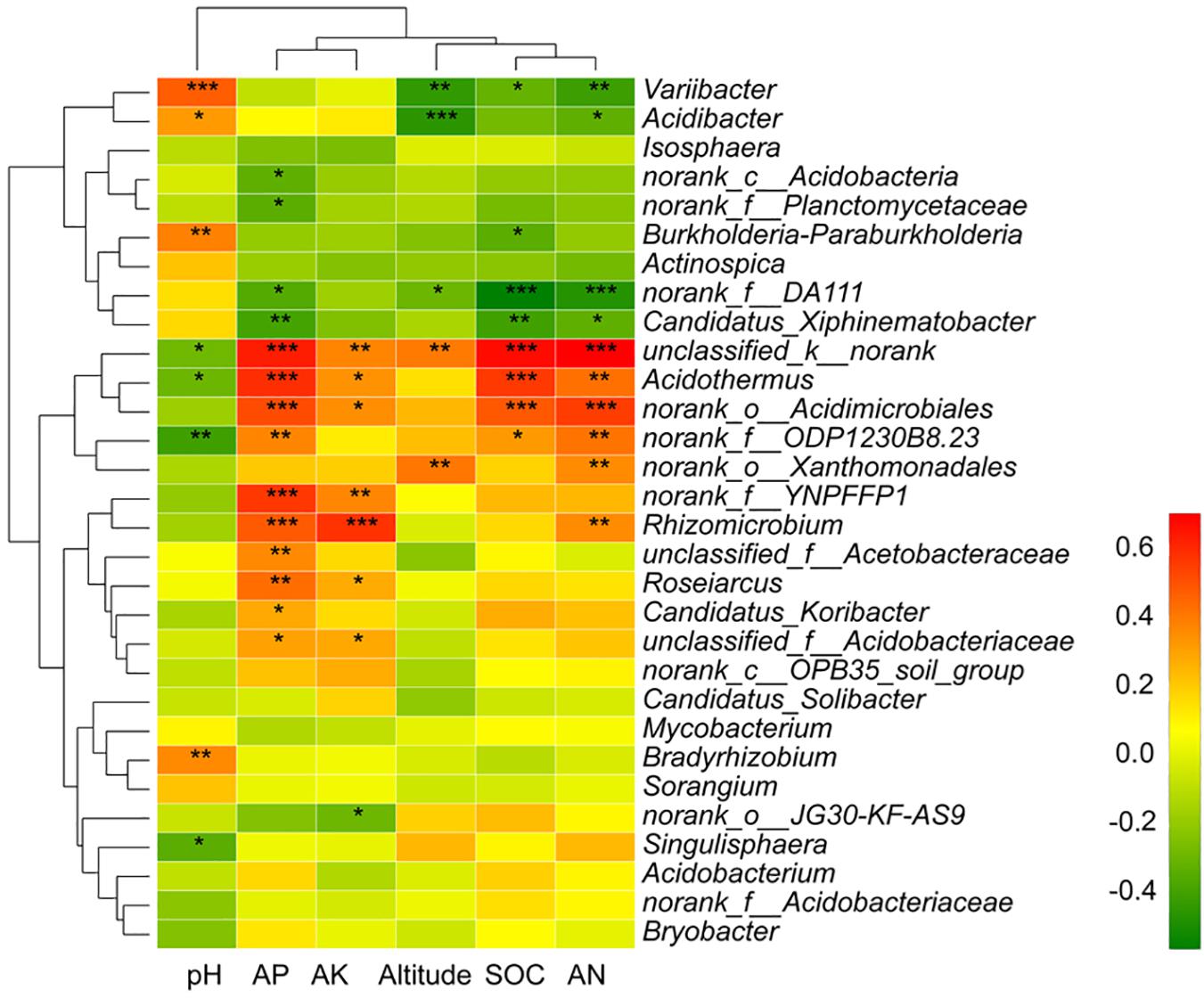
Figure 8. The Spearman correlation of the top 30 genera and soil/site properties. Significant differences by *p < 0.05; **p < 0.01 and ***p < 0.001.
Functional Predicted in Mycosphere and Bulk Soil
Using the Kyoto Encyclopedia of Genes and Genomes ortholog pathways (Oh et al., 2016), the KEGG functions of the identified bacteria were determined to be significantly (p < 0.05) affected by the mycosphere and bulk soil (Figure 9). The results showed that some functional traits, such as two-component system, bacterial chemotaxis, bacterial secretion system, tyrosine metabolism, biosynthesis of unsaturated fatty acids, ascorbate and aldarate metabolism, and metabolism of cofactors and vitamins, were significantly increase in mycosphere soil (p < 0.05) (Figure 9). When compared with bulk soil, valine, leucine, and isoleucine biosynthesis, ribosome biogenesis, homologous recombination, glycolysis/gluconeogenesis, and lysine biosynthesis were significantly (p < 0.05) lower in mycosphere soil (Figure 9).
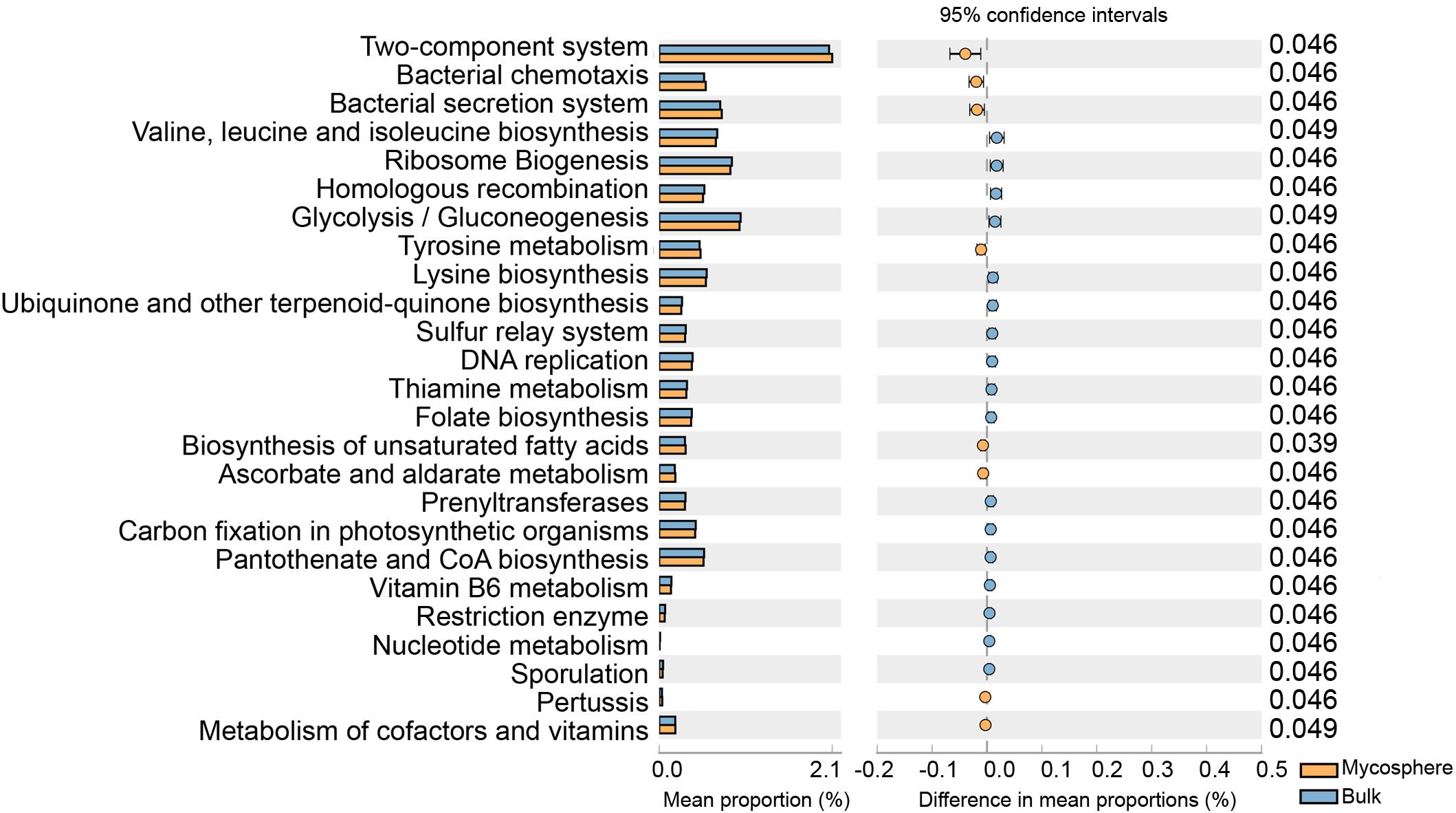
Figure 9. Comparison of the Kyoto Encyclopedia of Genes and Genomes function between mycosphere and bulk soil.
Discussion
Keystone Species and Ecological Functions
A considerable proportion (∼96%) of the coverage sequences is annotated to bacterial members (Figure 1), indicating that sequencing can be used to analyze the changes of the bacterial community structure in soil samples. Consistent with most of the earlier fungi research (Gryndler et al., 2000; Warmink and van Elsas, 2008; Oh et al., 2016), we found that, for most sites, bacterial diversity in the mycosphere soil was significantly lower than that in bulk soil. As seen in the R. griseocarnosa mycosphere soil (Figure 2), low bacterial diversity may be a common feature of the environment in which mycelium dominates (Gryndler et al., 2000). Compared to the bulk soil, Laccaria mycosphere bacterial diversity was significantly (p < 0.05) reduced on R2A agar analyses (Warmink and van Elsas, 2008). The bacterial diversity of Tricholoma matsutake dominant soil was significantly (p < 0.05) lower than T. matsutake minor soil (Oh et al., 2016). Olsson et al. (1996) demonstrated that ectomycorrhizal hyphae decreased the activity of bacteria in the soil. Therefore, it suggests that the variation of bacterial diversity might reflect the change of R. griseocarnosa population.
Proteobacteria, Acidobacteria, Actinobacteria, and Chloroflexi were the dominant bacterial communities in the soil (Figure 3), with an overall relative abundance higher than 86%. Proteobacteria, Planctomycetes, and Verrucomicrobia were significantly higher in the mycosphere soil, while Chloroflexi, Firmicutes, Cyanobacteria, Saccharibacteria, and Gemmatimonadetes were significantly lower. In some soil samples, the content of Acidobacteria in mycosphere soil was significantly (p < 0.05) higher than that in bulk soil (Supplementary Table S3).
Proteobacteria are naturally abundant in soil environments; thus, the increased richness found in the mycosphere soil could be the result of a positive effect of R. griseocarnosa because of its fast growth rate and its ability to use the major of root carbon substrates (Lauber et al., 2009). Proteobacteria increased richness might be stimulated by the higher nutritional status of soil in the mycosphere (Torsvik and Øvreås, 2002). Moreover, the dominance of Proteobacteria in hyphae (Cho et al., 2003), fruit bodies (Barbieri et al., 2010; Pent et al., 2017), and mycorrhizal roots (Poole et al., 2001; Frey-Klett et al., 2007) may be a result of the increased carbon content of these fungal-growing soils. Burke et al. (2006) described Acidobacterium as a MHB. Studies have shown that these Proteobacteria and Acidobacteria are physiologically and ecologically close, and both favor similar ecological niches in the rhizosphere soil (Singh et al., 2007; Kielak et al., 2016). Planctomycetes and Verrucomicrobia were significantly higher in plant rhizosphere soil (Stafford et al., 2005; Zul et al., 2007; Nunes da Rocha et al., 2009), and they seem to have a strong rhizospheric capacity functionally, but their role in the rhizospheric process remains to be proven.
Bacterial communities displayed distinct structures in the mycosphere and bulk soils (Figure 4 and Supplementary Table S4). Burkholderia-Paraburkholderia, Mycobacterium, Roseiarcus, Candidatus_Xiphinematobacter, Sorangium, Acidobacterium, and Singulisphaera were more abundant in the mycosphere soil than in the bulk soil samples (Figure 4 and Supplementary Table S4). The Proteobacteria genera Bradyrhizobium, Burkholderia-Paraburkholderia, and Roseiarcus are found in fungi-associated bacterial communities (Pent et al., 2017). For example, Burkholderia (Nguyen and Bruns, 2015) is known to be a mycorrhiza helper bacterium that promotes the growth and colonization of mycorrhizae. Kataoka et al. (2008) demonstrated that Burkholderia spp. and Bradyrhizobium spp. from ectomycorrhizal short roots with Russula and Suillus. Burkholderia spp. are well known as nitrogen-fixing bacteria (Timonen and Hurek, 2006). In recent years, many Burkholderia were reclassified as Paraburkholderia or Caballeronia (Sawana et al., 2014). For example, Burkholderia phenazinium and Burkholderia sordidicola were moved to the genus Paraburkholderia (Sawana et al., 2014), which are found in the mycorrhizosphere of Pinus muricata (Nguyen and Bruns, 2015). There is evidence that Burkholderia preferentially associates with mycorrhizal and that its strains can spread to the root tip (Poole et al., 2001). The members of the genus Burkholderia occur simultaneously with fungal taxa (Stopnisek et al., 2015), and the co-occurring might be due to Burkholderia’s ability to migrate with the growing hyphae (Nazir et al., 2012). Mycobacterium has nitrogen fixation functions (Rilling et al., 2018) and can provide nitrogen for the growth of R. griseocarnosa. Sorangium has rich xylan-degrading enzymes that can degrade biological macromolecules, cellulose, hemicellulose, and xylan (Tamaru et al., 2010), which is beneficial for increased mushroom productivity (Zhou et al., 2017). Singulisphaera, as an acidophilus, is also found in the rhizosphere soil of Boletus edulis (Mediavilla et al., 2019). Acidobacterium was significantly higher in plant rhizosphere soil (Oh et al., 2012; Yang et al., 2012), but their role remains to be proven in the rhizospheric process. It is indicated that Burkholderia-Paraburkholderia, Mycobacterium, Roseiarcus, Acidobacterium, Sorangium, and Singulisphaera were MHB of R. griseocarnosa. Although the functions of Candidatus Xiphinematobacter are unknown, it is possible that Candidatus Xiphinematobacter may be a MHB of R. griseocarnosa. These bacteria may play important roles in the growth of R. griseocarnosa.
Determinants of Bacterial Communities in Soil
The growth environment of the mycelium (ectomycorrhizal and mycosphere) affects both biological and abiotic factors in the soil ecosystem (Boersma et al., 2010; Kluber et al., 2010; Trappe et al., 2012). Through the study of fungi and bacteria in the mycosphere soil of T. matsutake, the results showed that the microbial diversity, community structure, and bacterial function in different geographical locations were similar (Oh et al., 2016). The diversity and community structure of mycosphere soil bacteria of Agaricus sinodeliciosus were different in different regions, but they all contained several main taxa (Zhou et al., 2017). R. griseocarnosa can co-exist with host tree species such as Betulaceae, Fagaceae, Pinaceae, and Tiliaceae to form ectomycorrhiza (Yu et al., 2020), but the symbiosis mechanism is still unclear (Yu et al., 2020), so we mainly studied the relationship between R. griseocarnosa and soil bacteria. There is growing evidence that root secretions regulate the relationship between mushrooms and soil microorganisms (Poole et al., 2001; Oh et al., 2016; Pent et al., 2017).
Russula griseocarnosa mycosphere has a high AN content in mycosphere soil (Table 1). Increased nitrogen supply can stimulate Russula to produce more spores and colonize more oak seedling roots (Avis et al., 2003). Soil pH and AN were significantly higher than most of the mycosphere soil samples (Supplementary Table S2). It was inferred that the main impact factors of R. griseocarnosa growth were pH and AN; moreover, previous research has found that pH significantly affects the soil’s bacterial community diversity (Fierer and Jackson, 2006; Rousk et al., 2010; Pent et al., 2017). Singh et al. (2008) found that fungal mycorrhizosphere and bacterial assemblage were affected by the soil pH. Here, the selected study locations had an acidic soil with pH values ranging from 3.99 to 4.55. Previous research showed that the changes in soil microbial community structures were closely related to soil chemistry (Cao et al., 2016). Several soil characteristics (e.g., nutrient availability and organic carbon) are directly or indirectly associated with soil pH, which may contribute to changes in the bacterial community structure (Rousk et al., 2010). Studies have found that higher (Singh et al., 2014) and medium (Meng et al., 2012; Siles and Margesin, 2016) elevations increase bacterial diversity, which is consistent with our findings that medium elevations increase bacterial diversity. The host plants and plant individuals have less of an effect on the diversity of soil rhizosphere bacteria, which is consistent with a previous study (Pivato et al., 2009).
Bacterial Function
Our study analyzed whether the bacterial communities of the mycosphere and bulk soils produce distinct functional profiles, thus linking R. griseocarnosa to specific functions of the bacterial soil. Our results indicated that mycospheres and bulk soils were functionally distinct. Mycosphere soils had an increase in the two-component system, bacterial chemotaxis, bacterial secretion system, tyrosine metabolism, biosynthesis of unsaturated fatty acids, ascorbate and aldarate metabolism, and metabolism of cofactors and vitamins (p < 0.05) (Figure 9). Pseudomonas can promote the growth of Agaricus bisporus, and the autophagy compounds secreted by A. bisporus can be degraded by Pseudomonas (Chen et al., 2013). Root exudates contain carbohydrates, amino acids, fatty acids, and vitamins, serve as a substrate for mycosphere microorganisms, and provide an important carbon source for soil microbes, thus contributing to the enrichment of the soil microbial community (Bais et al., 2006; Michielse et al., 2012). The increase of nutritional metabolism indicates that these bacteria prefer R. griseocarnosa mycosphere soil because it is easier to acquire nutrients (Oh et al., 2016). Although there are limitations in the interpretation of functional predictions, we have identified functions that have potentially positive impacts on R. griseocarnosa. Future research can address these functions to elucidate the dynamics among microorganisms in the R. griseocarnosa mycosphere soil.
The core functional genes in the mycosphere are not limited to a specific taxon (Yan et al., 2017). The relative abundance of some functional genes in the mycosphere was higher than in bulk soil, indicating that these functional traits were selected by the mycosphere. Although the mechanisms for the functional selection and its consequences in the mycosphere are unclear, our study provides valuable information to better understand the overly complex process of microbial community combinations in the mycosphere soil.
Conclusion
In conclusion, we identified a suitable environment for R. griseocarnosa growth by comparing the physicochemical properties, bacterial diversity, and community structure of mycosphere and bulk soils. 16S rRNA sequencing showed that the bacterial community composition in the mycosphere was significantly different from that of bulk soils. Further analysis showed that R. griseocarnosa growth caused a change in the microbial community structure. Growth of R. griseocarnosa reduces the diversity and abundance of soil bacterial communities. Among the soil variables, altitude and pH displayed significant contributions in bacterial community structure and diversity properties in all geographical sites under study. Soil pH and AN were the main factors contributing to R. griseocarnosa growth. We identified several dominant bacteria genera, including Mycobacterium, Roseiarcus, Candidatus_Xiphinematobacter, Sorangium, Acidobacterium, and Singulisphaera in the mycosphere that may improve R. griseocarnosa growth. In the functional analysis, we identified functional modules related to bacterial nutrient metabolism in the R. griseocarnosa mycosphere soil. The mycosphere soil is a complex environment, and our study shows that multiple symbiotic relationships between microbes and R. griseocarnosa might decrease bacterial diversity. Moreover, it suggests that the fruiting body formation of R. griseocarnosa may be affected not only by the host plants but also by the bacterial community in the mycosphere soil. Therefore, the application of management measures to improve soil properties, including the use of N fertilizer and microbial fertilizer containing MHB, may promote the conservation, propagation, and sustainable utilization of R. griseocarnosa.
Data Availability Statement
The Illumina sequencing raw reads were deposited into the NCBI BioProject: PRJNA553654 within GenBank. The SRA accession of raw reads number is SUB5929895.
Author Contributions
FY, J-FL, and JS participated in study design, sample collection, and statistical analyses. J-KL and S-KW conducted molecular biology experiments. FY drafted the manuscript. J-FL improved the manuscript.
Funding
This study was supported by the National Non-profit Institute Research Fund of CAF (CAFYBB2017ZB001), the National Natural Science Foundation of China (Nos. 31770657 and 31570544), and the Science and Technology Project of Guangdong Province (2017B020205002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00347/full#supplementary-material
Footnotes
References
Allison, S. D., Hanson, C. A., and Treseder, K. K. (2007). Nitrogen fertilization reduces diversity and alters community structure of active fungi in boreal ecosystems. Soil Biol. Biochem. 39, 1878–1887. doi: 10.1016/j.soilbio.2007.02.001 doi: 10.1016/j.soilbio.2007.02.001
Antony-Babu, S., Deveau, A., Van Nostrand, J. D., Zhou, J., Le Tacon, F., Robin, C., et al. (2013). Black truffle-associated bacterial communities during the development and maturation of Tuber melanosporum ascocarps and putative functional roles. Environ. Microbiol. 16, 2831–2847. doi: 10.1111/1462-2920.12294
Aspray, T. J., Frey-Klett, P., Jones, J. E., Whipps, J. M., Garbaye, J., and Bending, G. D. (2006). Mycorrhization helper bacteria: a case of specificity for altering ectomycorrhiza architecture but not ectomycorrhiza formation. Mycorrhiza 16, 533–541. doi: 10.1007/s00572-006-0068-3
Avis, P. G., McLaughlin, D. J., Dentinger, B. C., and Reich, P. B. (2003). Long-term increase in nitrogen supply alters above- and below-ground ectomycorrhizal communities and increases the dominance of Russula spp. in a temperate oak savanna. New Phytol. 160, 239–253. doi: 10.1046/j.1469-8137.2003.00865.x
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Barbieri, E., Ceccaroli, P., Saltarelli, R., Guidi, C., Potenza, L., Basaglia, M., et al. (2010). New evidence for nitrogen fixation within the Italian white truffle Tuber magnatum. Fungal Biol. 114, 936–942. doi: 10.1016/j.funbio.2010.09.001
Boer, W., de, Folman, L. B., Summerbell, R. C., and Boddy, L. (2005). Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29, 795–811. doi: 10.1016/j.femsre.2004.11.005
Boersma, F. G. H., Otten, R., Warmink, J. A., Nazir, R., and van Elsas, J. D. (2010). Selection of Variovorax paradoxus-like bacteria in the mycosphere and the role of fungal-released compounds. Soil Biol. Biochem. 42, 2137–2145. doi: 10.1016/j.soilbio.2010.08.009
Brulé, C., Frey-Klett, P., Pierrat, J. C., Courrier, S., Gérard, F., Lemoine, M. C., et al. (2001). Survival in the soil of the ectomycorrhizal fungus Laccaria bicolor and the effects of a mycorrhiza helper Pseudomonas fluorescens. Soil Biol. Biochem. 33, 1683–1694. doi: 10.1016/s0038-0717(01)00090-6
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Burke, D. J., Kretzer, A. M., Rygiewicz, P. T., and Topa, M. A. (2006). Soil bacterial diversity in a loblolly pine plantation: influence of ectomycorrhizas and fertilization. FEMS Microbiol. Ecol. 57, 409–419. doi: 10.1111/j.1574-6941.2006.00125.x
Cao, H., Chen, R., Wang, L., Jiang, L., Yang, F., Zheng, S., et al. (2016). Soil pH, total phosphorus, climate and distance are the major factors influencing microbial activity at a regional spatial scale. Sci. Rep. 6:25815. doi: 10.1038/srep25815
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, S., Qiu, C., Huang, T., Zhou, W., Qi, Y., Gao, Y., et al. (2013). Effect of 1-aminocyclopropane-1-carboxylic acid deaminase producing bacteria on the hyphal growth and primordium initiation of Agaricus bisporus. Fungal Ecol. 6, 110–118. doi: 10.1016/j.funeco.2012.08.003
Chen, X. H., Xia, L. X., Zhou, H. B., and Qiu, G. Z. (2010). Chemical Composition and Antioxidant Activities of Russula griseocarnosa sp. nov. J. Agric. Food Chem. 58, 6966–6971. doi: 10.1021/jf1011775
Cho, Y. S., Kim, J. S., Crowley, D. E., and Cho, B. G. (2003). Growth promotion of the edible fungus Pleurotus ostreatus by fluorescent pseudomonads. FEMS Microbiol. Lett. 218, 271–276. doi: 10.1016/s0378-1097(02)01144-8
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2013). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Delacre, M., Lakens, D., and Leys, C. (2017). Why psychologists should by default use welch’s t-test instead of student’s t-test. Int. Rev. Soc. Psychol. 30, 92–101. doi: 10.5334/irsp.82
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631. doi: 10.1073/pnas.0507535103
Frey-Klett, P., Burlinson, P., Deveau, A., Barret, M., Tarkka, M., and Sarniguet, A. (2011). Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 75, 583–609. doi: 10.1128/mmbr.00020-11
Frey-Klett, P., Garbaye, J., and Tarkka, M. (2007). The mycorrhiza helper bacteria revisited. New Phytol. 176, 22–36. doi: 10.1111/j.1469-8137.2007.02191.x
Garbeva, P., van Veen, J. A., and van Elsas, J. D. (2004). Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 42, 243–270. doi: 10.1146/annurev.phyto.42.012604.135455
Giomaro, G. M., Sisti, D., and Zambonelli, A. (2005). “Cultivation of edible ectomycorrhizal fungi by in vitro mycorrhizal synthesis,” in Vitro Culture of Mycorrhizas. Soil Biology, Vol. 4, eds S. Declerck, J. A. Fortin, and D. G. Strullu (Berlin: Springer), 253–267. doi: 10.1007/3-540-27331-x_14
Griffiths, R. I., Whiteley, A. S., O’Donnell, A. G., and Bailey, M. J. (2000). Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microb. 66, 5488–5491. doi: 10.1128/aem.66.12.5488-5491.2000
Gryndler, M., Hrselova, H., and Striteska, D. (2000). Effect of soil bacteria on hyphal growth of the arbuscular mycorrhizal fungus glomus claroideum. Folia Microbiol. 45, 545–551. doi: 10.1007/bf02818724
Hall, I. R., Yun, W., and Amicucci, A. (2003). Cultivation of edible ectomycorrhizal mushrooms. Trends Biotechnol. 21, 433–438. doi: 10.1016/s0167-7799(03)00204-x
Halsey, J. A., de Cássia Pereira e Silva, M., and Andreote, F. D. (2016). Bacterial selection by mycospheres of Atlantic Rainforest mushrooms. Anton. Leeuw. 109, 1353–1365. doi: 10.1007/s10482-016-0734-1
Kataoka, R., Taniguchi, T., Ooshima, H., and Futai, K. (2008). Comparison of the bacterial communities established on the mycorrhizae formed on Pinus thunbergii root tips by eight species of fungi. Plant Soil. 304, 267–275. doi: 10.1007/s11104-008-9548-x
Kielak, A. M., Barreto, C. C., Kowalchuk, G. A., van Veen, J. A., and Kuramae, E. E. (2016). The ecology of acidobacteria: moving beyond genes and genomes. Front. Microbiol. 7:744. doi: 10.3389/fmicb.2016.00744
Kluber, L. A., Tinnesand, K. M., Caldwell, B. A., Dunham, S. M., Yarwood, R. R., Bottomley, P. J., et al. (2010). Ectomycorrhizal mats alter forest soil biogeochemistry. Soil Biol. Biochem. 42, 1607–1613. doi: 10.1016/j.soilbio.2010.06.001
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil ph as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/aem.00335-09
Leyval, C., and Berthelin, J. (1991). Weathering of a mica by roots and rhizospheric microorganisms of pine. Soil Sci. Soc. Am. J. 55:1009. doi: 10.2136/sssaj1991.03615995005500040020x
Liu, P., Wang, X. H., Li, J. G., Qin, W., Xiao, C. Z., Zhao, X., et al. (2015). Pyrosequencing reveals fungal communities in the rhizosphere of Xinjiang jujube. Biomed Res. Int. 2015, 1–8. doi: 10.1155/2015/972481
Liu, Y., Zhang, J., and Meng, Z. (2018). Purification, characterization and anti-tumor activities of polysaccharides extracted from wild Russula griseocarnosa. Int. J. Biol. Macromol. 109, 1054–1060. doi: 10.1016/j.ijbiomac.2017.11.093
Lladó, S., López-Mondéjar, R., and Baldrian, P. (2017). Forest soil bacteria: diversity, involvement in ecosystem processes, and response to global change. Microbiol. Mol. Biol. Rev. 81, e63–e16. doi: 10.1128/mmbr.00063-16
Mantel Tests (1967). The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220.
Martin, F., Kohler, A., and Duplessis, S. (2007). Living in harmony in the wood underground: ectomycorrhizal genomics. Curr. Opin. Plant Biol. 10, 204–210. doi: 10.1016/j.pbi.2007.01.006
Mediavilla, O., Geml, J., Olaizola, J., Oria-de-Rueda, J. A., Baldrian, P., and Martín-Pinto, P. (2019). Effect of forest fire prevention treatments on bacterial communities associated with productive Boletus edulis sites. Microb. Biotechnol. 12, 1188–1198. doi: 10.1111/1751-7915.13395
Meng, H., Li, K., Nie, M., Wan, J. R., Quan, Z. X., Fang, C. M., et al. (2012). Responses of bacterial and fungal communities to an elevation gradient in a subtropical montane forest of China. Appl. Microbiol. Biot. 97, 2219–2230. doi: 10.1007/s00253-012-4063-7
Michielse, C. B., Reijnen, L., Olivain, C., Alabouvette, C., and Rep, M. (2012). Degradation of aromatic compounds through the β-ketoadipate pathway is required for pathogenicity of the tomato wilt pathogen Fusarium oxysporum f. sp. lycopersici. Mol. Plant Pathol. 13, 1089–1100. doi: 10.1111/j.1364-3703.2012.00818.x
Ming, T., Li, J., Huo, P., Wei, Y., and Chen, X. (2014). Analysis of free amino acids in russula griseocarnosa harvested at different stages of maturity using iTRAQ® -LC-MS/MS. Food Anal. Methods 7, 1816–1823. doi: 10.1007/s12161-014-9817-7
Mori, H., Maruyama, F., Kato, H., Toyoda, A., Dozono, A., Ohtsubo, Y., et al. (2014). Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res. 21, 217–227. doi: 10.1093/dnares/dst052
Munsch, P., Meyer, J. M., Marttinen, N., Gardan, L., Christen, R., and Alatossava, T. (2002). Pseudomonas costantinii sp. nov., another causal agent of brown blotch disease, isolated from cultivated mushroom sporophores in Finland. Int. J. Syst. Evol. Microbiol. 52, 1973–1983. doi: 10.1099/00207713-52-6-1973
Nazir, R., Hansen, M. A., Sorensen, S., and van Elsas, J. D. (2012). Draft genome sequence of the soil bacterium Burkholderia terrae strain BS001, which interacts with fungal surface structures. J. Bacteriol. 194, 4480–4481. doi: 10.1128/jb.00725-12
Nelson, D. W., and Sommers, L. E. (1996). Total carbon, organic carbon, and organic matter. Methods Soil Analy. Part 3 Chem. Methods 34, 961–1010. doi: 10.2136/sssabookser5.3.c34
Nguyen, N. H., and Bruns, T. D. (2015). The microbiome of Pinus muricata ectomycorrhizae: community assemblages, fungal species effects, and burkholderia as important bacteria in multipartnered symbioses. Microb. Ecol. 69, 914–921. doi: 10.1007/s00248-015-0574-y
Noble, R., Dobrovin-Pennington, A., Hobbs, P. J., Pederby, J., and Rodger, A. (2009). Volatile C8 compounds and pseudomonads influence primordium formation of Agaricus bisporus. Mycologia 101, 583–591. doi: 10.3852/07-194
Nunes da Rocha, U., Van Overbeek, L., and Van Elsas, J. D. (2009). Exploration of hitherto-uncultured bacteria from the rhizosphere. FEMS Microbiol. Ecol. 69, 313–328. doi: 10.1111/j.1574-6941.2009.00702.x
Oh, S. Y., Fong, J. J., Park, M. S., and Lim, Y. W. (2016). Distinctive feature of microbial communities and bacterial functional profiles in Tricholoma matsutake dominant soil. PLoS One 11:e0168573. doi: 10.1371/journal.pone.0168573
Oh, S. Y., and Lim, Y. W. (2018). Root-associated bacteria influencing mycelial growth of Tricholoma matsutake (pine mushroom). J. Microbiol. 56, 399–407. doi: 10.1007/s12275-018-7491-y
Oh, Y. M., Kim, M., Lee-Cruz, L., Lai-Hoe, A., Go, R., Ainuddin, N., et al. (2012). Distinctive bacterial communities in the rhizoplane of four tropical tree species. Microb. Ecol. 64, 1018–1027. doi: 10.1007/s00248-012-0082-2
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2017). Vegan: Community Ecology Package. R Package Version 2.4-3.
Olsson, P. A., Chalot, M., Bååth, E., Finlay, R. D., and Söderström, B. (1996). Ectomycorrhizal mycelia reduce bacterial activity in a sandy soil. FEMS Microbiol. Ecol. 21, 77–86. doi: 10.1016/0168-6496(96)00042-6
Parks, D. H., and Beiko, R. G. (2010). Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26, 715–721. doi: 10.1093/bioinformatics/btq041
Pent, M., Põldmaa, K., and Bahram, M. (2017). Bacterial communities in boreal forest mushrooms are shaped both by soil parameters and host identity. Front. Microbiol. 8:836. doi: 10.3389/fmicb.2017.00836
Pivato, B., Offre, P., Marchelli, S., Barbonaglia, B., Mougel, C., Lemanceau, P., et al. (2009). Bacterial effects on arbuscular mycorrhizal fungi and mycorrhiza development as influenced by the bacteria, fungi, and host plant. Mycorrhiza 19, 81–90. doi: 10.1007/s00572-008-0205-2
Poole, E. J., Bending, G. D., Whipps, J. M., and Read, D. J. (2001). Bacteria associated with Pinus sylvestris-Lactarius rufus ectomycorrhizas and their effects on mycorrhiza formation in vitro. New Phytol. 151, 743–751. doi: 10.1046/j.0028-646x.2001.00219.x
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Re. 41, D590–D596. doi: 10.1093/nar/gks1219
Retamal-Salgado, J., Hirzel, J., Walter, I., and Matus, I. (2017). Bioabsorption and bioaccumulation of cadmium in the straw and grain of Maize (Zea mays L.) in growing soils contaminated with cadmium in different environment. Int. J. Env. Res. Pub. Health. 14:1399. doi: 10.3390/ijerph14111399
Riedlinger, J., Schrey, S. D., Tarkka, M. T., Hampp, R., Kapur, M., and Fiedler, H. P. (2006). Auxofuran, a novel metabolite that stimulates the growth of fly agaric, is produced by the mycorrhiza helper bacterium streptomyces strain AcH 505. Appl. Environ. Microbiol. 72, 3550–3557. doi: 10.1128/aem.72.5.3550-3557.2006
Rilling, J. I., Acuña, J. J., Sadowsky, M. J., and Jorquera, M. A. (2018). Putative nitrogen-fixing bacteria associated with the rhizosphere and root endosphere of wheat plants grown in an andisol from southern chile. Front. Microbiol. 9:32881. doi: 10.3389/fmicb.2018.02710
Rousk, J., Bååth, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Sawana, A., Adeolu, M., and Gupta, R. S. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 5:429. doi: 10.3389/fgene.2014.00429
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Siles, J. A., and Margesin, R. (2016). Abundance and diversity of bacterial, archaeal, and fungal communities along an altitudinal gradient in alpine forest soils: what are the driving factors? Microb. Ecol. 72, 207–220. doi: 10.1007/s00248-016-0748-2
Singh, B. K., Munro, S., Potts, J. M., and Millard, P. (2007). Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl. Soil Ecol. 36, 147–155. doi: 10.1016/j.apsoil.2007.01.004
Singh, B. K., Nunan, N., Ridgway, K. P., McNicol, J., Young, J. P. W., Daniell, T. J., et al. (2008). Relationship between assemblages of mycorrhizal fungi and bacteria on grass roots. Environ. Microbiol. 10, 534–541. doi: 10.1111/j.1462-2920.2007.01474.x
Singh, D., Lee-Cruz, L., Kim, W.-S., Kerfahi, D., Chun, J. H., and Adams, J. M. (2014). Strong elevational trends in soil bacterial community composition on Mt. Halla, South Korea. Soil Biol. Biochem. 68, 140–149. doi: 10.1016/j.soilbio.2013.09.027
Stafford, W. H. L., Baker, G. C., Brown, S. A., Burton, S. G., and Cowan, D. A. (2005). Bacterial diversity in the rhizosphere of Proteaceae species. Environ. Microbiol. 7, 1755–1768. doi: 10.1111/j.1462-2920.2005.00929.x
Stopnisek, N., Zühlke, D., Carlier, A., Barberán, A., Fierer, N., Becher, D., et al. (2015). Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 10, 253–264. doi: 10.1038/ismej.2015.73
Storey, J. D. (2007). The optimal discovery procedure: a new approach to simultaneous significance testing. J. R. Stat. Soc. B. 69, 347–368. doi: 10.1111/j.1467-9868.2007.005592.x
Tamaru, Y., Miyake, H., Kuroda, K., Ueda, M., and Doi, R. H. (2010). Comparative genomics of the mesophilic cellulosome-producing clostridium cellulovorans and its application to biofuel production via consolidated bioprocessing. Environ. Technol. 31, 889–903. doi: 10.1080/09593330.2010.490856
Timonen, S., and Hurek, T. (2006). Characterization of culturable bacterial populations associating with Pinus sylvestris – Suillus bovinus mycorrhizospheres. Can. J. Microbiol. 52, 769–778. doi: 10.1139/w06-016
Torsvik, V., and Øvreås, L. (2002). Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5, 240–245. doi: 10.1016/s1369-5274(02)00324-7
Trappe, M. J., Cromack, K., Caldwell, B. A., Griffiths, R. P., and Trappe, J. M. (2012). Diversity of Mat-forming fungi in relation to soil properties, disturbance, and forest ecotype at crater Lake National Park, Oregon, USA. Diversity 4, 196–223. doi: 10.3390/d4020196
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., and Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
Warmink, J. A., Nazir, R., and van Elsas, J. D. (2009). Universal and species-specific bacterial “fungiphiles” in the mycospheres of different basidiomycetous fungi. Environ. Microbiol. 11, 300–312. doi: 10.1111/j.1462-2920.2008.01767.x
Warmink, J. A., and van Elsas, J. D. (2008). Selection of bacterial populations in the mycosphere of Laccaria proxima: is type III secretion involved? ISME J. 2, 887–900. doi: 10.1038/ismej.2008.41
Wu, J., He, Z. L., Wei, W. X., O’Donnell, A. G., and Syers, J. K. (2000). Quantifying microbial biomass phosphorus in acid soils. Biol. Fert. Soils 32, 500–507. doi: 10.1007/s003740000284
Yan, Y., Kuramae, E. E., de Hollander, M., Klinkhamer, P. G. L., and van Veen, J. A. (2017). Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J. 11, 56–66. doi: 10.1038/ismej.2016.108
Yang, T., Liu, G., Li, Y., Zhu, S., Zou, A., Qi, J., et al. (2012). Rhizosphere microbial communities and organic acids secreted by aluminum-tolerant and aluminum-sensitive soybean in acid soil. Biol. Fert. Soils 48, 97–108. doi: 10.1007/s00374-011-0608-7
Ye, J., Joseph, S. D., Ji, M., Nielsen, S., Mitchell, D. R. G., Donne, S., et al. (2017). Chemolithotrophic processes in the bacterial communities on the surface of mineral-enriched biochars. ISME J. 11, 1087–1101. doi: 10.1038/ismej.2016.187
Young, L. S., Chu, J. N., Hameed, A., and Young, C. C. (2013). Cultivable mushroom growth-promoting bacteria and their impact on Agaricus blazei productivity. Pesqui. Agropecu. Bras. 48, 636–644. doi: 10.1590/s0100-204x2013000600009
Yu, F., Song, J., Liang, J. F., Wang, S. K., and Lu, J. K. (2020). Whole genome sequencing and genome annotation of the wild edible mushroom, Russula griseocarnosa. Genomics 112, 603–614. doi: 10.1016/j.ygeno.2019.04.012
Yuan, Y., Liu, Y., Liu, M., Chen, Q., Jiao, Y., Liu, Y., et al. (2017). Optimization extraction and bioactivities of polysaccharide from wild Russula griseocarnosa. Saudi Pharm. J. 25, 523–530. doi: 10.1016/j.jsps.2017.04.018
Yun, Y. B., Park, S. W., Cha, J. S., and Kim, Y. K. (2013). Biological characterization of various strains of Pseudomonas tolaasii that causes brown blotch disease. J. Korean Soc. Appl. Biol. Chem. 56, 41–45. doi: 10.1007/s13765-012-2242-y
Zhao, J., Zhang, R., Xue, C., Xun, W., Sun, L., Xu, Y., et al. (2014). Pyrosequencing reveals contrasting soil bacterial diversity and community structure of two main winter wheat cropping systems in China. Microb. Ecol. 67, 443–453. doi: 10.1007/s00248-013-0322-0
Zhou, J., Bai, X., and Zhao, R. (2017). Microbial communities in the native habitats of Agaricus sinodeliciosus from xinjiang province revealed by amplicon sequencing. Sci. Rep. 7:15719. doi: 10.1038/s41598-017-16082-1
Keywords: Russula griseocarnosa, mycosphere, functional diversity, Miseq sequencing, soil bacterial community
Citation: Yu F, Liang J-F, Song J, Wang S-K and Lu J-K (2020) Bacterial Community Selection of Russula griseocarnosa Mycosphere Soil. Front. Microbiol. 11:347. doi: 10.3389/fmicb.2020.00347
Received: 14 August 2019; Accepted: 17 February 2020;
Published: 25 March 2020.
Edited by:
Anna Maria Pirttilä, University of Oulu, FinlandReviewed by:
Christophe Calvayrac, Université de Perpignan Via Domitia, FranceRam Prasad, Mahatma Gandhi Central University, India
Risto Kasanen, University of Helsinki, Finland
Copyright © 2020 Yu, Liang, Song, Wang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun-Feng Liang, jfliang2000@163.com
 Fei Yu
Fei Yu Jun-Feng Liang
Jun-Feng Liang Jie Song
Jie Song Sheng-Kun Wang
Sheng-Kun Wang Jun-Kun Lu
Jun-Kun Lu