- 1Microbiology Immunology Department, Faculty of Biology, University of Bucharest, Bucharest, Romania
- 2The Research Institute of the University of Bucharest, Bucharest, Romania
- 3Department of Life, Medical and Agricultural Sciences, Biological Sciences Section, Academy of Romanian Scientists, Bucharest, Romania
- 4Faculty of Medicine and Biological Sciences, Stefan cel Mare University of Suceava, Suceava, Romania
- 5Microbiology Department, Suceava Emergency County Hospital, Suceava, Romania
- 6Cellular and Molecular Pathology Department, Stefan S. Nicolau Institute of Virology, Bucharest, Romania
- 7Institute of Biology of Romanian Academy, Bucharest, Romania
- 8Department of Biochemistry and Molecular Biology, Faculty of Biology, University of Bucharest, Bucharest, Romania
- 9ENT Department, University of Medicine and Pharmacy Carol Davila and Coltea Clinical Hospital, Bucharest, Romania
- 10DANUBIUS Department, National Institute of Research and Development for Biological Sciences, Bucharest, Romania
Mammalians sense antigenic messages from infectious agents that penetrate the respiratory and digestive epithelium, as well as signals from damaged host cells through membrane and cytosolic receptors. The transduction of these signals triggers a personalized response, depending on the nature of the stimulus and the host’s genetics, physiological condition, and comorbidities. Interferons (IFNs) are the primary effectors of the innate immune response, and their synthesis is activated in most cells within a few hours after pathogen invasion. IFNs are primarily synthesized in infected cells, but their anti-infective effect is extended to the neighboring cells by autocrine and paracrine action. The emergence of the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) pandemic in 2019 was a stark reminder of the potential threat posed by newly emerging viruses. This pandemic has also triggered an overwhelming influx of research studies aiming to unveil the mechanisms of protective versus pathogenic host immune responses induced by SARS‐CoV‐2. The purpose of this review is to describe the role of IFNs as vital players in the battle against SARS‐CoV-2 infection. We will briefly characterize and classify IFNs, present the inductors of IFN synthesis, their sensors, and signaling pathways, and then discuss the role of IFNs in controlling the evolution of SARS-CoV-2 infection and its clinical outcome. Finally, we will present the perspectives and controversies regarding the prophylactic and therapeutic potential of IFNs in SARS-CoV-2 infection.
1 Introduction
Viral infections still cause significant morbidity and mortality globally, despite vaccination, which has majorly contributed to decreasing their burden or even eradicating them. The balance between the viral host conquering strategies and the host immune response dictates the clinical outcome of a viral infection. Therefore, elucidating the intimate mechanisms of the antiviral immune response is essential for developing new preventive, diagnosis, and therapeutic strategies (1).
Viral aggression is initially countered by the innate defense mechanisms located at the level of the epithelial barrier, involving genetic, chemical, and biological factors (2, 3). If the virus surpasses the innate host defenses and starts replicating, the host will respond by triggering an inflammatory response mediated by effector molecules and cells of the innate immune system mobilized from the local blood vessels at the site of infection (4). If the innate immune response fails to clear the infection, even after being augmented by increased production of effector molecules and cells, an adaptive immune response will be initiated (2, 5).
The inflammatory response typically involves four key components: inflammatory inducers, sensors that detect these inducers, inflammatory mediators induced by the sensors, and the target tissues affected by these mediators. The damaged tissue macrophages will recognize, bind, and phagocytose the viral pathogen through their surface receptors and activate an inflammatory response, leading to the accumulation of humoral (plasma proteins such as complement components) and cellular (neutrophils, macrophages, and dendritic cells – DCs) effectors of the innate immunity. Viral, bacterial, and fungal infectious agents show a wide range of associated antigens named “pathogen-associated molecular pattern” (PAMP), quickly recognized by epithelial cells, macrophages, and resident DCs, through innate immunity sensors called “pattern recognition receptors” (PRR) (6). PRR sensors recognize a broad pattern of PAMP antigens and play an essential role in developing innate and adaptative immune responses. The best-known PRR sensors that PAMP activates are classified in 4 families: (i) Toll-like receptors (TLR), (ii) retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) that includes RIG I, melanoma-associated differentiating protein 5 (MDSA5), and Laboratory of Genetics and Physiology 2 (LGP 2) (iii) NOD-like receptors family (NLR) (NOD- nucleotide-binding oligomerization domain), and (iv) C-type lectin sensors (7, 8). After recognizing the danger signals, the cell activates the molecular effectors mediating the aggression response.
Interferons (IFNs) are essential players in the inflammatory innate response, significantly impacting cellular, tissue, and overall physiological functions (9, 10). IFNs, as suggested by their name, were initially discovered for their remarkable ability to interfere with and counteract viral infections in the 1950s (11). The production of IFNs is triggered by a wide array of pattern recognition receptors (PRRs) (12). Once IFNs are produced, they initiate an intrinsic antiviral state within the cells that detect them, characterized by the activation of interferon-stimulated genes (ISGs), which are instrumental in antiviral defense (13).
The emergence of the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) pandemic in 2019 was a stark reminder of the potential threat posed by newly emerging viruses. This pandemic has also triggered an overwhelming influx of research studies to understand the different mechanisms involved in the SARS‐CoV‐2 triggered protective or harmful immune responses. Given the critical role of IFNs in controlling viral infections, these cytokines have emerged as vital players in the battle against SARS‐CoV-2 infection (14–27). While IFNs are essential for controlling SARS-CoV-2, they can also potentially contribute to developing severe COVID-19 clinical forms (28–30). This variety and complexity of immune responses is caused by the virus’s ability to evade or manipulate the IFN-mediated host responses, which might not be perfectly tuned to combat this novel pathogen (31).
The purpose of this review is to describe the role of IFNs as vital players in the battle against SARS‐CoV-2 infection. We will briefly characterize and classify IFNs, present the inductors of IFN synthesis, their sensors, and signaling pathways, and then discuss the role of IFNs in controlling the evolution of SARS-CoV-2 infection and its clinical outcome. Finally, we will present the perspectives and controversies regarding the prophylactic and therapeutic potential of IFNs in SARS-CoV-2 infection.
2 General characterization and classification of IFNs
IFNs are one of the six groups of cytokines (i.e., interleukins-ILs, tumor necrosis factors-TNFs, chemokines, transforming growth factors-TGFs, hemopoietic growth factors, and IFNs) produced by the host cells primarily in response to viral pathogens. Four types of IFNs, noted I (IFNα with 13 subtypes, IFNβ, IFNϵ, IFNκ, IFNω found in bovines and humans, IFNδ found in pig, IFNτ, IFNς or limitin and trofoblastic IFNτ in ruminants), II (IFN-γ), III (IFN-λ1-4 sharing similarities with the IL-10 family cytokines) and IV were described until now (18, 32–46).
Human IFNs are proteins of 130-170 amino acids with a molecular weight of 20 - 100 kDa, active at temperatures generally lower than 56-60°C and at different pH values (47–51). IFNα and IFNβ (are acid-stable, while IFNγ is acid-labile (13, 52). IFNs exhibit different activities, including the induction of an antiviral state, inhibition of normal and tumor cell multiplication, cell differentiation regulation, and immune function regulation by modulating the expression of major histocompatibility complex (MHC) molecules (48, 53). Type I and III IFNs are the first responders when a viral threat is detected and can be produced by most cell types. In contrast, type II IFNs are primarily generated by specialized immune cells like NK, B, TCD8+, and TCD4+ cells, participating mainly in allergic responses, host defense against intracellular pathogens, and the control of tumor growth. This distinction highlights the diverse roles and functions of different IFNs (54–56). Type IV IFN (IFN-υ or IFN-U), along with its receptors (IFN-υR1 and IL10R2), has been recently discovered in jawed vertebrates (45).
3 Inductors of IFN synthesis
In the absence of activating signals, normal cells do not produce detectable levels of IFN. IFN synthesis is triggered by different infective agents: viruses, mycoplasma, rickettsia and chlamydia, ds RNA, synthetic polymers, mannans, and metabolic activators (31, 57–59). The first demonstrated, and most important stimulators of IFN synthesis are the nucleoprotein complexes of ds RNA viruses, as demonstrated by Isaacs and Lindenmann (11). DNA viruses, except Poxviruses, are weak inducers, while the ssRNA genome induces IFN through their ds intermediates produced during replication (60). Inactivated viruses and those infecting non-permissive substrates are also IFN synthesis inducers (61). In mononuclear leukocytes, IFN synthesis is induced by the viral envelope glycoproteins (62).
The main classes of PAMP receptors are i) TLR sensors harboring a cytoplasmic domain with a sequence similar to Toll/IL1R (TIR) receptor and stimulating IFN I, IFN III, and other mediators involved in inflammatory and adaptative immune responses synthesis (37, 63–66); ii) NLR (NOD-like receptor) receptors are cytosolic proteins that control this compartment for intracellular aggressor signals; iii) RLR sensors (retinoic acid-inducible gene I) are a vast cytosolic helicase protein family acting as the most important sensors for ds RNA, ss RNA, and DNA viruses (67–71); iii) cGAS (Cyclic GMP-AMP synthase), secondary endogenous messenger sensing the cytosolic DNA and consequently synthesizing the second messenger, cGAMP, further binding and activating STING (stimulator of IFN genes), which on its turn, activates TBK1 kinase that induces the STING-dependent IFNb synthesis through the IRF 3 (IFN regulatory factor 3) (72–74); iv) Type C lectin sensors (e.g., mannose receptors) are carbohydrate structures associated with macrophage, CD, and Langerhans cells membrane that recognize carbohydrate surface structures of bacteria, viruses, and parasites (7, 75); v) ALR sensor (Absent In Melanoma 2 = AIM-2-Like Receptor), detected in odontoblast layer (76), recognizes cytosolic and pathogen DNA and is inductive of IFN I synthesis (77, 78).
ISG can be activated by the common mechanisms of the other PRR sensors: activator phosphorylation and nuclear translocation of cytoplasmic factors IRF3, IRF7, and NF-kB (79).
Activation of IFN synthesis is a complex process consisting of the following: (i) viral antigen recognition mediated by macrophages, DCs, and Langerhans epithelial cells receptors, (ii) signal transduction through the adaptor kinase pathway, (iii) phosphorylation and nuclear translocation of transcription factors IRF3/IRF7 (IFN regulatory factor), and (iv) stimulation of ISG transcription (80–83). IRF and NF-kB activation trigger two antiviral programs: (i) cell defense by induction of antiviral status mediated by IFN I, which in its turn activates leukocytes, especially neutrophils and IFN III, and (ii) activation of an extended ISG genes series (48).
4 IFNs receptors and signal transduction pathways
The IFN molecule has two domains with relatively constant amino acid sequences: the N-terminal domain that forms the B (binding) situs on the cell receptor, and the other A (activity) in the C-terminal domain that seems to modulate receptor binding and mediate the adaptative and innate immune responses (50). Their main role is the induction of viral multiplication inhibitor protein synthesis. IFN receptor distribution is strictly regulated, dictating the cell capacity to answer IFNs.
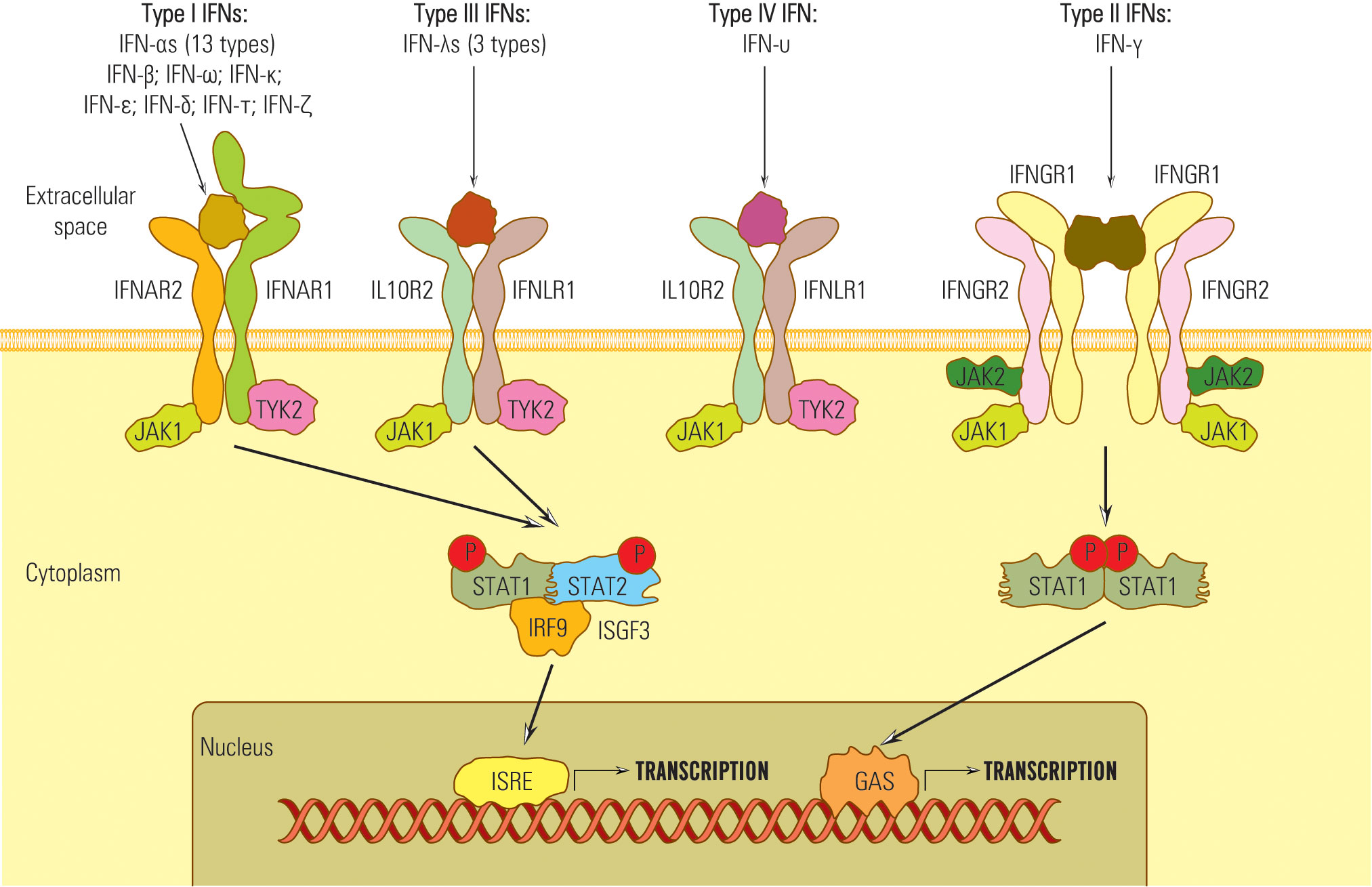
Despite their diverse amino-acid homologies, all type I IFNs transmit signals through a common heterodimeric receptor consisting of low-affinity (IFNAR1) and high-affinity (IFNAR2) receptor components (84) (Figure 1).
In order to transmit signals via the JAK/STAT pathway, both IFNAR1 and IFNAR2 receptors form associations with tyrosine kinase 2 (Tyk2) (in the case of IFNAR1) and Janus kinase 1 (JAK1) in the case of IFNAR2 (85, 86) (Figure 1). These kinase associations are essential for the phosphorylation of the receptor and the recruitment of STAT (signal transducer and activator of transcription) proteins to the receptor complex (87, 88).
In contrast to type I and type II IFNs, which have distinct receptors, the IFNλ share a common receptor, IFNLR1, with another group of cytokines, including IL-10, IL-22, and IL-26 (IL10RB) (34). All type III IFNs transmit signals through a common heterodimeric receptor consisting of two subunits: IFNLR1, also referred to as IL28Rα, and IL10Rβ (84, 89–91). The signaling complex employed by type III IFNs comprises four transmembrane-spanning receptors, which include two copies of each of the high-affinity receptor IFNGR1 and low-affinity receptor IFNGR2 (84) (Figure 1). The activation of the JAK/STAT signaling pathway in response to IFNλ binding to its receptor complex is facilitated by JAK1, associated with IFNLR1, and respectively by Tyk2 linked with IL10RB (39).
Therefore, IFNs I, II, and III signals are essentially transduced by JAK/STAT pathway, but other pathways might be involved, which explains the diversity of IFNs biological effects (50, 92, 93) (Figure 2).
Notably, type IV IFN shares a C-terminal sequence with type I IFN and utilizes the same signal-transduction receptor, IL10R2, as type III IFN. This suggests a common ancestral origin for type IV IFN and type I/III IFNs (45, 94).
Upon activation, STAT kinases attract IRF9 and form the heterotrimer complex STAT1-STAT 2-IRF-9 known as ISGF 3 (IFN stimulated gene factor 3) (95). ISGF 3 complex migrates into the nucleus, activates ISRE (IFN stimulated response element) belonging to the ISG gene promoter, and starts transcription (96).
The STAT combinations that bind ISRE are STAT 1-STAT 2, STAT 2-STAT 3-6, or STAT 2 homodimers that generate diversity among IFN-activated genes and effects, e.g., STAT 1 recruitment induces pro-inflammatory, antiproliferative, and proapoptotic effects; STAT 3,4,5 stimulate T lymphocyte proliferation (48, 56, 97, 98).
IFN III activates the STAT 1-STAT 2 pathway and regulates gene transcription through ISRE or by GAS elements (56, 89, 99) (Figure 1). STAT complexes that do not attract IRF 9 bind to the GAS site (IFN-γ activated site) and activate ISG promoters. For example, IFN-γ does not induce ISGF3 complex formation and, consequently, does not activate genes with only ISRE in their promoters (53, 100).
Besides the JAK/STAT pathway, IFN I, II, and III, by JAK kinases, can activate other non-STAT pathways: MAPK (mitogen-activated protein kinase), crucial in the activation of antiproliferative and antiviral genes depending on IFNI and PI3K pathway (35, 101).
Some genes can be induced in the absence of IFN. Some are activated by IRF1, IRF3, IRF 7, NF-kB, or IL-1 (56, 102).
5 Antiviral IFN activity
Even though IFNs are not effector molecules, they induce an antiviral state that depends on extracellular IFN level, cell type, viral virulence, functional status of immune effectors, and activity of the molecular complex of PRR sensors, signal transduction pathways, and IFN codifying genes.
The in vitro IFN synthesis is activated in the first 2-3 hours and reaches a maximum in 16 hours after macrophage infection by the influenza virus (103). Type I IFN, produced by the infected cells simultaneously with the viral multiplication cell cycle, is released from cells immediately after synthesis, and diffuses toward the non-infected cells.
IFN I and III mediated antiviral state is a consequence of two mechanisms (104). First, in most virus-cell systems, viral multiplication inhibition results from interference with mRNA translation: IFN inhibits the binding of ribosomes to viral messengers. Secondly, in infected cells, IFN activates the expression of hundreds of ISG cluster genes that mediate pleiotropic effects, counting antiviral, antiangiogenic, immunomodulatory, cell cycle inhibitory, apoptotic, and antitumor activities (105–108). Genes activated by IFN I and IFN III codify a variety of antiviral effectors (109–111). ISG activation leads to mRNA synthesis, but some hundreds of ISG encode non-coding RNA. Other molecules, besides IFNs, could directly activate some ISGs, such as IRF, NF-kB, or IL-1 (112). Others have a basic transcription level, the IFNs only exhibiting a stimulatory or repressor effect. The response to different viruses is individual (109).
The antiviral protective effect induced by IFN is present in all stages of the multiplication cycle: cell entry, decapsidation, transcription, translation and replication of viral RNA, assembly, and release of virions. Infections with IFN-sensitive viruses are more severe in laboratory animals previously injected with anti-IFN I or actinomycin D (a specific protein synthesis inhibitor) (113, 114).
The most essential antiviral effectors with IFN I induced synthesis are RNA-dependent protein kinase (PKR) and 2’ 5’ oligo-adenosine synthetase (OAS), specific adenosine deaminase for RNA (ADAR), the product of gene 56 GTP ase, Mx, etc. (54, 97, 115). The antiviral effect was proved especially for PKR and for 2’-5’-OAS/RNase L system. Both are constitutively expressed in normal cells as inactive latent forms. The minimum mRNA level for these cells is stimulated by IFN I (109, 116–118). PKR is one of the four kinases from mammalian cells, the most critical antiproliferative and antiviral effector induced by IFN I (119). The enzyme is mainly found in the cytoplasm (80%) associated with ribosomes and is more phosphorylated than the nuclear one. In the absence of inductors, PKR is synthesized in a lower amount, non-phosphorylated, monomeric, and inactive. After binding of IFN I to membrane receptor or/and in the presence of dsRNA or influenza nucleoprotein, PKR level increases, dimerizes, self-phosphorylates, and phosphorylates the eukaryotic protein synthesis factor (eIF2α) and inhibits viral mRNA translation (67, 120, 121). The 2’-5’oligoadenylat synthetases (OAS) bind dsDNA and are activated by conformational change. The major role of 2’-5’OA is the activation of RNase L, constitutively synthesized and detectable in all animal cells, that, in its turn, destroys viral RNA and blocks viral multiplication, and also induces apoptosis (6, 122–124). The 2’-5’OAS enzyme could exhibit antiviral activity also by RNase L independent pathways.
Other antiviral effectors are: i) protein 56 (p56) that binds the 3e initiating factor of the eukaryotic cell (eIF3e), inhibits mRNA translation initiation, and thus activates apoptosis (125, 126); ii) DICER protein from the dsRNA binding family (DRBP- =ds binding protein) is involved in gene inactivation mediated by RNA interference (67).
IFN a/b has an indirect protective effect by decreasing the permeability of the digestive and respiratory tract mucosa and consolidating the blood-brain barrier (116). It has been shown that neurons, astrocytes, and microglia synthesize IFN I with an anti-pathogenic protective effect (127, 128). Also, IFN stimulates the major histocompatibility complex molecules (MHC) from class I and II and their co-stimulators, which are encoded by ISG genes, on the surface of antigen-presenting cells (APC) (100). IFN I also stimulate chemokine-mediated APC migration (48).
IFN I protects cells from the viral cytopathic effect but does not completely eliminate the infection. Antiviral protective status IFN lasts several days and can be reinduced (127).
Inadequate IFN I synthesis amplifies the pathogenic effects of acute and persistent viral infection by immunosuppressive effects and aberrant inflammatory reactions (129). IFN I chronic synthesis can be associated with clinical manifestations: infectious diseases, neoplasia, chronic inflammation, and autoimmune diseases. Mutations in genes codifying synthesis for IFN, IFN receptors, or activator signal transduction pathways might generate a risk of developing viral or bacterial infections (130). Some ISG genes, such as adenosine deaminase (ADAR), can stimulate certain viruses’ multiplication (131, 132).
IFN γ is a pleiotropic cytokine with contradictory effects reported in viral infections, depending on the virus, the intensity of the innate and adaptative immune response, pro-inflammatory cytokine synthesis rate, and underlying pathology. It stimulates cellular immune response, protects against Mycobacterium tuberculosis, inhibits the multiplication of certain viruses, activates macrophages and reactive oxygen species (ROS) release, and induces macrophage polarization to M1 pro-inflammatory phenotype but maintains the M1/M2 ratio (55, 133). M1 macrophages have microbicidal activity and release pro-inflammatory cytokines (IL-1β, IL12, TNFα) and M2 IL-10 (anti-inflammatory) and TGF (tumor growth factor) (134). On the other hand, IFN-γ stimulates immunotolerance in chronic viral infection, induces pro-inflammatory cytokine synthesis in SARS-CoV-2 infection (cytokine storm), determines lesions of the lung epithelium, microvascular endothelium, ischemia, pulmonary fibrosis (18). Other authors reported the blocking of experimental liver fibrosis by IFN-γ (135).
Type III IFN regulates the activity for several gene sets and determines biological activities similar to IFN I, the only difference being that IFN III produces limited pro-inflammatory effects (136).
The recent discovery of the type IV IFN system and its antiviral functions still raises several important questions and needs future research to elucidate signaling networks and if the regulatory relationships of type IV IFNs with the other IFN types are synergistic, antagonistic, or independent. Secondly, while type IV IFN has demonstrated strong antiviral properties, it’s important to explore whether it also exerts regulatory functions against other types of pathogens, such as bacteria, fungi, parasites, etc., similar to the type I and II IFNs (94). As a cytokine, it’s important to explore the broader regulatory functions of type IV IFN in various immune cells, including lymphocytes, macrophages, DCs, thrombocytes, neutrophils, etc., and its impact on inflammation, phagocytosis, or other immune responses.
6 IFNs in SARS-CoV-2 infection
More than half of human infections are zoonotic, and of the 224 human infective viruses, 88% are zoonotic (137). However, only a few adapted to a human host and initiated pandemics in the last 200 years: H1N1 influenza virus (1918) from birds (138), swine variants of influenza A (2009 pandemics –pdm-09) (139), avian influenza A H7N9 (140); HIV from Macaccus (32.7 million deaths) (141); hepatitis C virus (unknown origin, infected more than 70 million people) (142); MERS from camels (143); SARS-CoV-2 associated with Civeta civetictis, bats and pangolin (144); Hendra and Nipah viruses (paramyxoviruses) spread by bats (145); encephalitis agents isolated from horse and pig (146). Other viruses with animal-to-human transmission belong to rodents: Hantavirus and Machupo arenaviruses, Lassa, and Junin. In most zoonotic infections, humans represent the final host (147).
IFN I is essential to eliminate the virus and curtail the immune response swiftly (90), being vital in regulating T effector cells, responsible for virus elimination, and for the differentiation of regulatory CD4+ T cells that produce the inhibitory cytokines like IL-10. Blocking IFN signaling during MERS-CoV infection in mice reduced the development of virus-specific CD4+ and CD8+ T cells (148). Studies on IFN I receptor knockout (IFNAR-/-) mice have shown that this can lead to increased pathology due to elevated production of proinflammatory cytokines triggered by the viral infection (149). Response of the target cells to specific cytokines, including IFN I, is regulated by IFN concentration, receptor expression, and viral mechanisms for counteracting the immune response. Chronic IFN I synthesis could have pathologic effects, such as stimulation of antigen presentation and activation of more lymphocyte clones, including self-reactive ones, that can initiate an autoimmune response. The reverse IFN effect synthesizes IL-10, a cytokine with immunosuppressive and pro-apoptotic effects (102).
Experimental data shows that respiratory epithelial cell (nose, oropharynx, nasopharynx, larynx, sinuses, conjunctiva) synthesis of IFN I seems to play a determinant role in the dynamic and severity of SARS-CoV-2 infection (150). The rate of transcription for IFN, AR1, JAK1, and TYK2, transducers of IFN synthesis activating signal (ISG), is higher in patients with mild or moderate infection associated with the increased plasma level of IFN, by contrast with a decrease in ISG expression mediated by inhibitor genes MX1, IFITM1, IFIT2 and decreased IFN I blood level in patients with severe infection (151, 152). Infection severity is associated with family deficiency of ISG or IFN I encoding genes and anti-IFN I autoantibodies (28, 153). Macrophages, DC, and keratinocytes produce IFN κ, and its protective antiviral functions are inhibited in SARS-CoV-1 infection (48). DC produces IFN ω and exhibits antiviral activity against SARS-CoV-2, specific antibodies detected in COVID-19 patients with severe pneumonia (153, 154).
However, while early IFN I production is crucial for an effective T cell response (reducing the SARS-CoV-2 density, severity, and duration of clinical infection), delayed IFN response can inhibit T cell proliferation and result in T cell exhaustion and death (155, 156). Thus, timing of IFN synthesis during infection is essential for stopping infection evolution.
Also, a high blood level of IFN I is not always a marker for viral protection. Longitudinal analysis of viral load for respiratory tract epithelium in critically ill patients with COVID is proportional with IFN-α, IFN-γ, and RNF level, proving that SARS-CoV-2 multiplication is not always regulated by IFN I (28, 157, 158).
The presence of lung injury in severe COVID-19 cases suggests a potential failure to activate immunosuppressive mechanisms promptly. Patients with more severe COVID-19 symptoms tend to have lower counts of regulatory T (Treg) cells influenced by IFNs (159). The decreasing number of Treg cells raises the hypothesis that dysregulated IFN responses elicited by SARS-CoV-2 may impact Treg cell generation during the recovery phase of COVID-19. Future studies should investigate the role of IFN dysregulation in shaping T cell responses and how it may, in turn, affect antibody responses since CD4+ T cell activation is crucial for B cell immunity. A deeper understanding of these interactions will provide valuable insights into the immune response during COVID-19 (160).
Existing comorbidities decreased the immune response and increased the pulmonary pathological process mediated by pro-inflammatory IL produced by macrophages. Pre-stimulated macrophages by external inflammatory factors had the same effect (28, 161, 162).
Non-adequate quantitative response or delayed IFN synthesis in SARS-CoV-2 infection induced the activation of many ISG genes with immunopathologic potential, such as overexpression of genes encoding pro-inflammatory cytokine synthesis, exacerbating the inflammatory reaction (163). In the plasma of critically ill COVID-19 patients, IFN I, IL 6, and TNF α had high concentrations, showing increased IFN I activity mainly from pDCs and neutrophils, that generate a cytokine storm (164, 165).
Regarding type II IFN, the results of the cross-sectional analysis conducted by Piater and collaborators in 142 infected patients show that in most COVID-19 patients, IFN-γ-mediated biochemical pathways were still strongly activated after 60 days. The authors observed that the ongoing activation of IFN-γ-mediated pathways might influence the further course of reconvalescence, and the continuous immune activation might go along with enhanced demand for nutrients like amino acids and vitamins (24). Mansoor and collaborators aimed to investigate the crosstalk between host immune response mediated by cytokines and the severity of SARS-CoV-2 infection by assessing cytokine expression in 136 infected patients. In this regard, the authors measured the expression levels of 12 genes encoding inflammatory, anti-inflammatory, and regulatory cytokines using QRT-PCR in hospitalized patients with severe infection and found that IFN-γ could be a potent marker of disease severity (22). Primorac and colleagues designed a study involving 303 participants who were tested for the analysis of IFN-γ concentration and the detection of human antibodies of the immunoglobulin class IgG against the S1 domain of the SARS-CoV-2 spike protein. The statistical analysis revealed a significant difference in the IFN-γ concentration between participants who had experienced reinfections and those who had not been infected. Participants who had not been infected or reinfected with SARS-CoV-2 after vaccination and before SARS-CoV-2 infection displayed a notably higher level of cellular immunity. Additionally, among individuals who had not received additional vaccination, those who had experienced infection or reinfection had significantly lower IFN-γ levels than uninfected participants. These findings suggest that cellular immunity, as measured by IFN-γ concentrations, has a lasting impact and is crucial in preventing infections and reinfections, especially in the context of emerging SARS-CoV-2 variants of concern (25).
Suzuki and collaborators conducted a study to investigate the potential of 71 humoral factors as predictive markers of COVID‐19 in 188 patients diagnosed with COVID‐19 using antigen or nucleic acid amplification tests. The authors showed that IFNλ3 predicted subsequent oxygen demand better than other humoral factors (e.g., CRP, LDH, lymphocyte fraction) in patients in the early phase of COVID‐19 without supplemental oxygen demand. IFNλ3 may effectively predict whether a patient with COVID‐19 will require medical intervention, such as oxygen supplementation, at an earlier point before the patient presents with respiratory failure. In conclusion, in patients with COVID-19 who do not require supplemental oxygen for the first week after the onset of the disease, the serum IFNλ3 level is a highly accurate predictor for the likelihood of needing oxygen support later (21). These findings hold significant importance in making early decisions regarding patient placement and initiating timely therapeutic interventions. Looking ahead, IFNλ3 could potentially serve as a valuable tool for enhancing the prognosis of COVID-19 patients while alleviating the strain on healthcare facilities.
A recent study conducted by Matic and collaborators determined that the presence of the most frequent functional single nucleotide polymorphisms (SNPs) of the two most important IFN-λs coding genes, namely IFNL3 and IFNL4, could alter the likelihood of SARS-CoV-2-infected patients to develop a more severe form of the disease. This clinical study involving 178 COVID-19 patients revealed that carriers of IFNL3 and IFNL4 minor alleles are less likely to progress from mild to moderate COVID-19, that is, to develop COVID-19-related pneumonia. Also, the authors observed that the likelihood of pneumonia development remained significantly associated with IFNL4 polymorphism, especially in females. These results suggest that IFNL4 rs12979860 and rs368234815 polymorphisms could predict the risk of COVID-19-related pneumonia development in females (23).
7 Viral strategies against IFNs
During the SARS-CoV-2 pandemic, the interest in understanding the mechanisms by which animals tolerate viral infections has increased. Rodents and bats have developed strategies for overcoming or deleting the immune response (166). Bats have an increased response to viral RNA and a decreased response to cytoplasmic DNA viruses because of the lack of genes codifying DNA cytoplasmic sensors. In humans, most viruses induce synthesis of IFN and ISG gene transcription stimulating factors, but the most virulent neutralize the IFN response. For most viruses, the gene expression inductor for IFN α/β is dsRNA. Many viruses generated strategies that prevent dsRNA exposure in the cytoplasm (167–169). In reoviruses, dsRNA remains inside the capsid during the whole viral cycle. Also, most replication intermediates are associated with viral proteins covering ds viral RNA regions; thus, only a small amount of dsRNA is exposed (170, 171).
In most cases, viruses inhibit the innate immune response by the action of nonstructural and structural proteins that inhibit transmission of IFN synthesis activating signals (172, 173). Most viruses encode molecules that destroy PKR, inhibit the interaction between PKR and ds RNA, and inhibit PKR phosphorylation (173–175). Poxviruses encode proteins that inhibit the host immune response by blocking IFN, TNF, IL-1 chemokines synthesis, and transduction of apoptosis activating signal (176). One of the mechanisms by which bats tolerate the SARS-CoV-2 infection is that ISG transcription is constitutively expressed at low levels, and regulatory genes are lacking. Inflammation is limited by inhibiting TNF-α expression associated with a decreased level of NLRP3 inflammasome activation. Bats response to viral infection is, in fact, anti-inflammatory, being associated with the expression of IL-10 (177, 178).
In the case of SARS-CoV-2, some structural proteins and most SARS-CoV-2 nonstructural proteins (nsp) are involved in counteracting the IFN synthesis activating signals by different pathways: (i) M protein can interact with the MDA5 sensor, or with MAVS adaptor protein or TBK1, part of the IFN I synthesis activating signal transduction chain, (ii) M protein induces ubiquitin and TBK1 degradation and blocks transduction of IFN synthesis activating signal (179), (iii) nsp1 inhibits cell mRNA translation and IFN synthesis (180, 181), (iv) ORF 9b protein inhibits the signaling pathway for RIG –I/MDA5-MAVS adaptor proteins TRIF and STING for TLR-3 TRIF signaling pathway as well as cGAS-ATING pathway, the sensor of cytosolic DNA by which IRF 3 phosphorylation and nuclear translocation are blocked (182), (v) the SARS-CoV-2 ARN-polymerase blocks IRF 3 nuclear translocation as well as ORF 6 protein (183), (vi) ns 3 and ns 5 proteases cleave viral polyproteins and also cell proteins, blocking the IFN receptors by direct action, (vii) N protein covers the genomic RNA, inhibits dsRNA recognition by RIG I, and inhibits phosphorylation of STAT 1, and STAT 2 kinases and their translocation in the nucleus (98, 184), (viii) nsp 6 and nsp 13 bind TBK1 and delete the IRF activation; ix) other ns proteins block the activator signal for STAT 1/STAT 2 phosphorylation and their translocation in the nucleus (98, 183), and (x) coronaviruses replicate in structures limited by membranes that protect viral RNA from cell sensors (Figure 3).
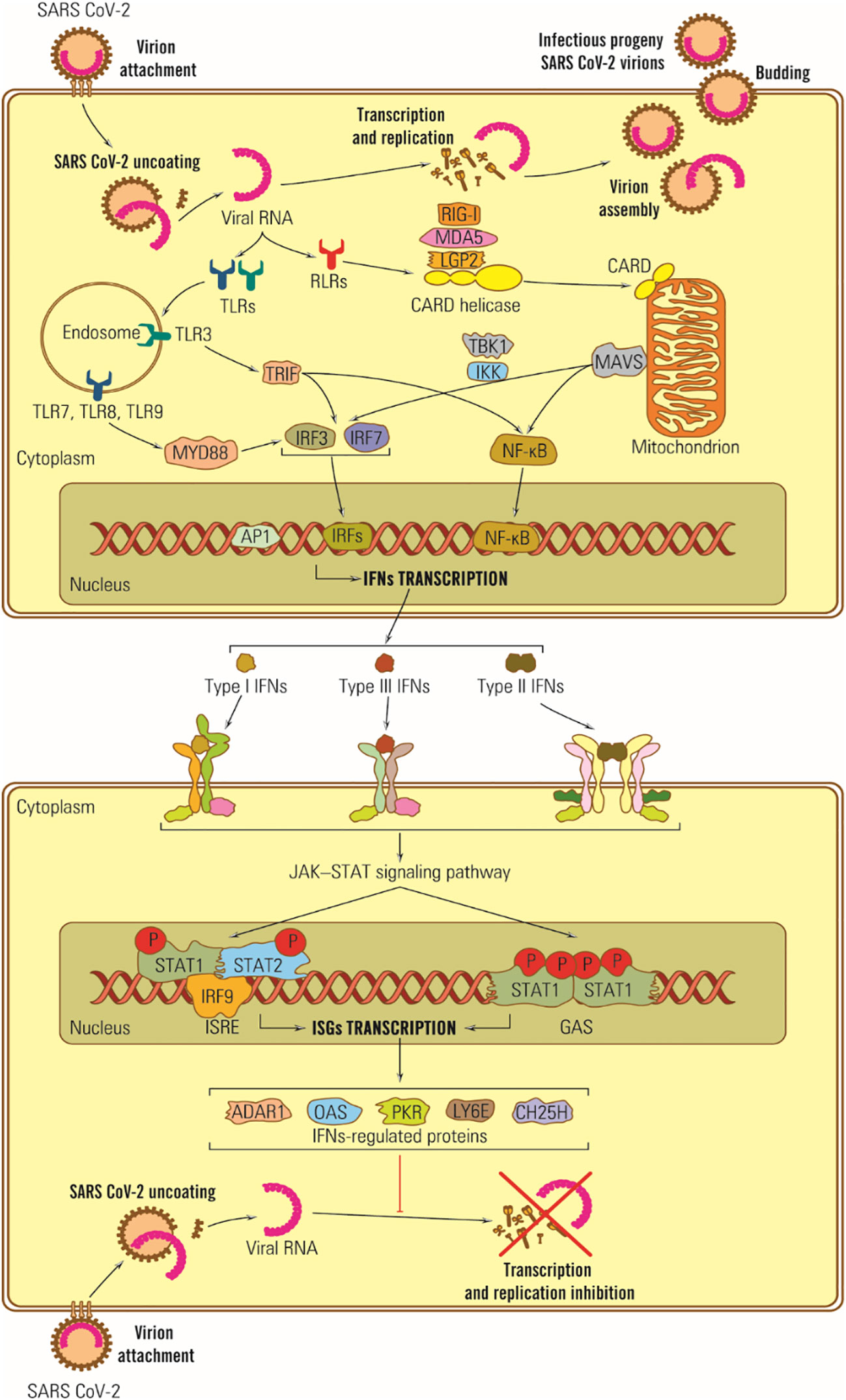
Figure 3 Schematic representation of IFN antiviral effects in SARS-CoV-2 infection. Viral RNA is recognized by TLR-3, -7 and -8 located on the endosomal membrane, and protein E is directly bound to TLR-2. Its dysfunction plays a vital role in the CNS response to SARS-CoV-2 infection, potentially triggering neurodegenerative lesions. Ds subgenomic RNA synthesized in mRNA transcription is recognized by RIG I and MDA-5 (185).
The ability of SARS-CoV-2 proteins to interfere with IFN I questions the use of the whole virus as a vaccine. On the other hand, vaccinating with specific SARS-CoV-2 proteins that lack IFN I regulatory activity, like the S-protein, may lead to enhanced IFN I production and a stronger immune response (186).
8 Potential of IFNs for the therapeutic management of COVID-19
The dysregulation of IFNs in COVID-19 highlights its importance in the disease’s development and its potential as a therapeutic target. IFNs administered prophylactically can create an antiviral state in cells, potentially preventing early-stage viral infections (151). In animal studies, early IFN treatment before viral replication peak was beneficial, while late administration hindered viral clearance and worsened immunopathology (155).
An experimental trial in 2020 demonstrated that daily IFN-α nasal drops, along with standard protective equipment, protected at-risk healthcare workers from COVID-19 for 28 days without adverse effects (NCT04320238).
Clinical evidence suggests that administering IFN I interferon early in SARS-CoV-2 infection is more effective in reducing disease severity and mortality compared to late administration (14, 20). The early treatment can probably speed up viral clearance, leading to quicker recovery and a decreased risk of severe illness. Moreover, early-stage IFN treatment may also play a role in reducing virus transmission (187).
Pandit and collaborators led a clinical trial to investigate the effectiveness and safety of pegylated interferon alfa-2b (PEG IFN-α2b) in 40 patients with moderate COVID-19. The primary endpoint was the improvement in clinical status on day 15, measured by the WHO 7-point ordinal scale. Overall, 19 subjects who received PEG IFN-α2b had improved clinically on day 15, and 80% had a negative RT-PCR result on day 7. Mild adverse events were reported for eleven subjects. This study highlights the significant improvement in clinical status on day 15 due to faster viral reduction due to the PEG IFN-α2b administration in moderate COVID-19 subjects (17). Levy and collaborators reported the safety of a single subcutaneous injection of Peg-IFN-α2a in two patients with inborn errors of the TLR-3 and IRF-7, affecting the production of type I IFNs and predisposing to severe COVID-19. Both patients reported a rapid decrease in the symptoms and signs present at admission following the administration of Peg-IFN-α2a, which suggests that these types of patients may benefit from the very early administration of type I IFN. Furthermore, this observation suggests that the earliest possible administration should be considered in trials of type I IFN-based regimens for treating SARS-CoV-2-infected patients at risk of developing severe disease (16).
Recently, Jhuti and collaborators reviewed eleven studies reporting the benefits of IFN therapy for the treatment of SARS-CoV-2 and, based on the findings, formulated several recommendations to be taken into account in IFN therapy: (i) standardized outcome measures, (ii) dedicated analyses by interferon type, (iii) analyses of effectiveness by disease stage, (iv) exploration of higher IFN dosage, and (v) cost analysis (19).
Although clinical studies combining IFNs with antivirals for SARS-CoV and MERS-CoV yielded inconclusive results due to variable administration timings and comorbidities (15), however, the most effective approach seems to be combining interferon with other repurposed antiretroviral drugs, such as lopinavir, ritonavir, and remdesivir (188, 189).
A recent finding has shown that that ACE2 is an IFN-stimulated gene in the human airway epithelial cells. This finding raises questions about whether prophylactic or therapeutic IFN administration might enhance SARS-CoV-2 entry and replication during disease progression (15).
Recent studies found that patients with COVID-19 exhibited an increase in the production of IFN-γ, even before the release of antibodies. As a result, this potent cytokine appears to be the primary mediator of innate and adaptive immune responses during SARS-CoV-2 infection and a potential candidate for therapeutical approaches (18, 190).
Myasnikov and collaborators conducted a randomized study including patients with moderate COVID-19 infection, aiming to assess the effect of subcutaneous administration of IFN-γ (500,000 IU, s/c, daily, once a day, for five days) in patients with viral pneumonia on the changes in vital signs and the duration of hospital stay. The study showed that IFN-y resulted in more favorable changes in the stabilization of vital signs, as well as in reduced length of fever and hospital stay by two days, which suggests a positive effect of this substance on the recovery processes in patients with moderate COVID-19. Notably, patients who received recombinant IFN-γ experienced no progression of respiratory failure and required no transfer to the intensive care unit. These results confirm the positive effect of IFN-y on the clinical stabilization and recovery rate of patients with community-acquired pneumonia and viral infections (191). In a study led by van Laarhoven and collaborators, five critically ill COVID-19 patients with severe defects in cellular immune responses, high SARS-CoV-2 viral RNA loads, and no respiratory improvement were treated with IFN-γ (100 μg subcutaneously, thrice weekly). Bronchial secretion was collected every 48 hours for routine diagnostic SARS-CoV-2 RT-PCR and viral culture. A rapid decline followed IFN-γ administration in SARS-CoV-2 load and positive-to-negative viral culture conversion. The results revealed no signs of hyperinflammation, allowing us to consider IFN-γ as adjuvant immunotherapy in a subset of immunocompromised COVID-19 patients (192).
Despite these promising results, in a clinical trial conducted by Roquilly and collaborators, the results indicated that interferon gamma-1b treatment (100 µg interferon gamma-1b every 48 hours for a period of 1 to 9 days) did not significantly reduce the incidence of disease or death in the first 28 days. In addition, the study was stopped due to safety concerns about interferon gamma-1b treatment (193). These divergent outcomes suggest that further research is needed to determine the efficacy of IFN-γ in both prophylaxis and treatment of COVID-19.
Research on the antiviral properties of interferon lambda (IFN-λ) in the context of intestinal infections has emphasized its enduring and non-inflammatory characteristics. As a result, numerous studies have delved into the prospect of utilizing IFN-λ in COVID-19.
In a study conducted by Feld and collaborators, the benefits of administering a single subcutaneous injection of pegylated IFN-λ were discussed, particularly in the treatment of mild to moderate COVID-19 within the first seven days of symptom onset or upon the first positive swab in asymptomatic cases (194). However, in another randomized, placebo-controlled study involving 120 patients with mild to moderate COVID-19, the subcutaneous administration of pegylated IFN-λ1 within 72 hours of diagnosis did not lead to a shorter duration of SARS-CoV-2 viral shedding nor did it result in symptom improvement (195). These divergent outcomes suggest that further research is needed to determine the efficacy of pegylated IFN-λ in both prophylaxis and treatment of COVID-19 due to the uncertainty surrounding its effects. Also, the exact timing for applying IFN-λ-based therapeutics could be crucial: it should be earlier to significantly reduce the viral load and thus decrease the overall severity of the disease.
More recently, Reis and collaborators conducted a randomized clinical trial (NCT04727424) aiming to investigate the efficacy of a single dose of pegylated IFN-λ in vaccinated adults with SARS-CoV-2 infection from Brazil and Canada. In total, 933 patients received pegylated IFN-λ(single subcutaneous injection, 180 μg), and 1018 received placebo (single injection or oral). This study revealed that the vaccine’s effectiveness remained consistent across different dominant virus variants, irrespective of the patient’s vaccination status. Among individuals with a high initial viral load, those who were administered pegylated IFN-λ showed a notable reduction in viral load by the seventh day compared to those who received a placebo. Furthermore, the incidence of adverse events was comparable in both groups, indicating the treatment’s safety. The authors concluded that among predominantly vaccinated outpatients with COVID-19, those who received a single dose of pegylated IFN-λ had a significantly lower likelihood of being hospitalized or visiting the emergency department than those who received a placebo (26).In addition, Santer and colleagues aimed to determine if a peripheral immune cell response to therapeutic administration of pegylated IFN-λ in vivo could be detected. In this context, the authors performed single-cell RNA sequencing (scRNAseq) on 9 patients receiving pegylated IFN-λ. ScRNAseq was performed to investigate the expression of the IFN-λ receptor (IFNLR1/IL10RB) and to detect in vivo interferon-stimulated gene responses in individual immune cell populations. After filtering for high-quality cells, the authors included 263,668 cells, 146,408 cells from pegylated IFN-λ-treated, and 117,260 from placebo patients. ScRNAseq confirmed in vivo responses to pegylated IFN-λ in specific peripheral immune cells, but the treatment did not alter virus-specific adaptive immune responses. The antiviral effects of pegylated IFN-λ were observed despite a delayed T-cell response in older patients at risk of more severe outcomes (196).
Ryoo and collaborators conducted a comprehensive analysis to assess the effectiveness and safety of IFN (systemic or inhaled IFN-α, -β, and -λ) treatment in patients with COVID-19, stratified by the severity of their clinical condition. The meta-analysis incorporated data from 11 clinical trials encompassing 6,124 patients. The collective findings from this analysis indicated that, compared to a placebo, IFN therapy did not yield significant improvements in reducing mortality at day 28 or preventing the progression to mechanical ventilation in COVID-19 patients. However, it did exhibit a noteworthy increase in the rate of hospital discharge on day 14 when compared to the control group. In summary, IFN therapy was considered safe but did not exhibit favorable outcomes regarding critical clinical endpoints in COVID-19 patients, particularly those with more than moderate disease severity. Importantly, IFN therapy did not appear to worsen outcomes in patients with severe COVID-19. The study suggests that future clinical trials should focus on assessing the clinical efficacy of IFN therapy in patients with mild COVID-19 or at an earlier stage of the disease (27).
Research should investigate whether type IV IFN can be harnessed as a therapeutic protein. It would be significant to understand its potential therapeutic applications and evaluate its safety and efficacy in clinical settings (94). Addressing these questions and conducting further research will enhance our understanding of the type IV IFN system and its potential roles in immune response and therapy.
Taken together, the current evidence suggests that future clinical trials should focus on assessing the clinical efficacy of IFN therapy in patients with different clinical forms of COVID-19, from mild to severe and at different stages of the disease (27).
9 Conclusions
Timely coordinated cellular and humoral innate and acquired immune responses dictate the progression and severity of viral infections. One of the main effectors of antiviral innate immunity are the IFNs, which exert their antiviral activity through direct and indirect effects. IFNs interact with specific cell receptors and trigger activation signals for hundreds of genes that determine a broad spectrum of effects: inducing the antiviral state, inhibiting cellular proliferation, apoptosis activation, and stimulating the immune response. However, delayed IFN I synthesis could have opposite effects by stimulating the pro-inflammatory cytokine release and amplifying the pathologic process. The ability of SARS-CoV-2 proteins to interfere with IFN I response could explain the evolution and progression of infection in different individuals and indicate the potential therapeutic benefits of IFN in COVID-19.
Interferon treatments offer multiple benefits, including easy administration initiated by healthcare providers, potential acceleration of viral clearance, and quicker clinical improvement, especially when administered early in SARS-CoV-2 infection. Additionally, interferon therapy is associated with minimal reported side effects like temporary nausea and digestive problems.
However, the results of the available studies are contradictory, and to enable patients to benefit more effectively from IFN’s therapeutic use, there is a pressing need for standardization of interventional studies. Also, a deeper understanding of the timing and dynamics of IFN responses during SARS-CoV-2 infections is crucial for informing IFN-related therapies and vaccine development.
Author contributions
GM: Writing – original draft, Writing – review & editing. MCC: Conceptualization, Writing – original draft, Writing – review & editing. RF: Writing – review & editing. CB: Writing – review & editing. LMD: Writing – review & editing. MC: Writing – review & editing, Figure design. R-EC: Writing – original draft, Writing – review & editing. RG: Writing – review & editing. SVB: Writing – review & editing. GB: Writing – review & editing. COV: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Romanian Executive Agency for Higher Education, Research, Development, and Innovation (https://uefiscdi.gov.ro/, accessed on 20 April 2023) research project FDI 0690/2023, “The core program within the National Research Development and Innovation Plan, 2022–2027”, carried out with the support of the Ministry of Research, Innovation and Digitalization (MCID), project no. 23020101, Contract no. 7N from 3 January 2023, and the “Analysis of the potential for sustainable use of vegetation specific to the Danube-Danube Delta-Black Sea system” project, awarded by the European Regional Development Fund through the Competitiveness Operational Program 2014–2020, contract no. 108630.The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Weber F. Antiviral innate immunity: introduction. Encyclopedia Virol (2021), 1:577–83. doi: 10.1016/B978-0-12-809633-8.21290-9
2. Janeway CA Jr, Travers P, Walport M, Shlomchik MJ. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science (2001). 884 p.
3. Chen X, Liu S, Goraya MU, Maarouf M, Huang S, Chen JL. Host immune response to influenza A virus infection. Front Immunol (2018) 9:320. doi: 10.3389/fimmu.2018.00320
4. Kany S, Vollrath JT, Relja B. Cytokines in inflammatory disease. Int J Mol Sci (2019) 20(23):6008. doi: 10.3390/ijms20236008
5. Marshall JS, Warrington R, Watson W, Kim HL. An introduction to immunology and immunopathology. Allergy Asthma Clin Immunol (2018) 14(2):49. doi: 10.1186/s13223-018-0278-1
6. Li LF, Yu J, Zhang Y, Yang Q, Li Y, Zhang L, et al. Interferon-inducible oligoadenylate synthetase-like protein acts as an antiviral effector against classical swine fever virus via the MDA5-mediated type I interferon-signaling pathway. J Virol (2017) 91(11):e01514–16. doi: 10.1128/JVI.01514-16
7. Drouin M, Saenz J, Chiffoleau E. C-type lectin-like receptors: head or tail in cell death immunity. Front Immunol (2020) 11:251. doi: 10.3389/fimmu.2020.00251
8. Alipor DS, Misraeidi M. Inborn errors in the LRR domain of Nod2 and their potential consequences on the function of the receptor. Cells (2021) 10(8):2031. doi: 10.3390/cells10082031
9. Li D, Wu M. Pattern recognition receptors in health and diseases. Signal Transduct Target Ther (2021) 6(1):291. doi: 10.1038/s41392-021-00687-0
10. Walker FC, Sridhar PR, Baldridge MT. Differential roles of interferons in innate responses to mucosal viral infections. Trends Immunol (2021) 42(11):1009–23. doi: 10.1016/j.it.2021.09.003
11. Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci (1957) 147(927):258–67. doi: 10.1098/rspb.1957.0048
12. Mertowska P, Smolak K, Mertowski S, Grywalska E. Immunomodulatory role of interferons in viral and bacterial infections. Int J Mol Sci (2023) 24(12):10115. doi: 10.3390/ijms241210115
13. Dowling JW, Forero A. Beyond good and evil: molecular mechanisms of type I and III IFN functions. J Immunol (2022) 208(2):247–56. doi: 10.4049/jimmunol.2100707
14. Davoudi-Monfared E, Rahmani H, Khalili H, Hajiabdolbaghi M, Salehi M, Abbasian L, et al. A randomized clinical trial of the efficacy and safety of interferon β-1a in treatment of severe COVID-19. Antimicrob Agents Chemother (2020) 64(9):e01061–20. doi: 10.1128/AAC.01061-20
15. Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell (2020) 181(5):1016–1035.e19. doi: 10.1016/j.cell.2020.04.035
16. Levy R, Bastard P, Lanternier F, Lecuit M, Zhang SY, Casanova JL. IFN-α2a therapy in two patients with inborn errors of TLR3 and IRF3 infected with SARS coV-2. J Clin Immunol (2021) 41(1):26–7. doi: 10.1007/s10875-020-00933-0
17. Pandit A, Bhalani N, Bhushan BLS, Koradia P, Gargiya S, Bhomia V, et al. Efficacy and safety of pegylated interferon alfa-2b in moderate COVID-19: A phase II, randomized, controlled, open-label study. Int J Infect Dis (2021) 105:516–21. doi: 10.1016/j.ijid.2021.03.015
18. Todorovic-Rakovic N, Whitfield JR. Between immunomodulation and immunotolerance: The role of IFNγ in SARS CoV-2 disease. Cytokine (2021) 146:155637. doi: 10.1016/j.cyto.2021.155637
19. Jhuti D, Rawat A, Guo CM, Wilson LA, Mills EJ, Forrest JI. Interferon treatments for SARS CoV-2: Challenges and opportunities. Infect Dis Ther (2022) 11(3):953–72. doi: 10.1007/s40121-022-00633-9
20. Salto-Alejandre S, Palacios-Baena ZR, Arribas JR, Berenguer J, Carratalà J, Jarrín I, et al. Impact of early interferon-β treatment on the prognosis of patients with COVID-19 in the first wave: a post hoc analysis from a multicenter cohort. BioMed Pharmacother. (2022) 146:112572. doi: 10.1016/j.biopha.2021.112572
21. Suzuki T, Iwamoto N, Tsuzuki S, Kakumoto Y, Suzuki M, Ashida S, et al. Interferon lambda 3 in the early phase of coronavirus disease-19 can predict oxygen requirement. Eur J Clin Invest. (2022) 52(9):e13808. doi: 10.1111/eci.13808
22. Mansoor S, Butt AR, Bibi A, Mushtaq S, Ullah I, Alshahrani F, et al. Expression of IFN-Gamma is significantly reduced during severity of covid-19 infection in hospitalized patients. PloS One (2023) 18(9):e0291332. doi: 10.1371/journal.pone.0291332
23. Matic S, Milovanovic D, Mijailovic Z, Djurdjevic P, Sazdanovic P, Stefanovic S, et al. It’s all about IFN-λ4: Protective role of IFNL4 polymorphism against COVID-19-related pneumonia in females. J Med Virol (2023) 95(10):e29152. doi: 10.1002/jmv.29152
24. Piater T, Gietl M, Hofer S, Gostner JM, Sahanic S, Tancevski I, et al. Persistent symptoms and IFN-γ-mediated pathways after COVID-19. J Pers Med (2023) 13(7):1055. doi: 10.3390/jpm13071055
25. Primorac D, Brlek P, Pavelić ES, Mešić J, Glavaš Weinberger D, Matišić V, et al. Importance of cellular immunity and IFN-γ Concentration in preventing SARS coV-2 infection and reinfection: A cohort study. Viruses. (2023) 15(3):792. doi: 10.3390/v15030792
26. Reis G, Moreira Silva EAS, Medeiros Silva DC, Thabane L, Campos VHS, Ferreira TS, et al. Early treatment with pegylated interferon lambda for Covid-19. N Engl J Med (2023) 388(6):518–28. doi: 10.1056/NEJMoa2209760
27. Ryoo S, Koh DH, Yu SY, Choi M, Huh K, Yeom JS, et al. Clinical efficacy and safety of interferon (Type I and Type III) therapy in patients with COVID-19: A systematic review and meta-analysis of randomized controlled trials. PloS One (2023) 18(3):e0272826. doi: 10.1371/journal.pone.0272826
28. Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature (2020) 584(7821):463–9. doi: 10.1038/s41586-020-2588-y
29. Dorgham K, Quentric P, Gokkaya M, Marot S, Parizot C, Sauce D, et al. Distinct cytokine profiles associated with COVID-19 severity and mortality. J Allergy Clin Immunol (2021) 147:2098–107. doi: 10.1016/j.jaci.2021.03.047
30. Galani IE, Rovina N, Lampropoulou V, Triantafyllia V, Manioudaki M, Pavlos E, et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat Immunol (2021) 22:32–40. doi: 10.1038/s41590-020-00840-x
31. Zhang S, Wang L, Cheng G. The battle between host and SARS-CoV-2: innate immunity and viral evasion strategies. Mol Ther (2022) 30:1869–84. doi: 10.1016/j.ymthe.2022.02.014
32. Weissenbach J, Chernajovsky Y, Zeevi M, Shulman L, Soreq H, Nir U, et al. Two interferon mRNAs in human fibroblasts: in vitro translation and Escherichia coli cloning studies. Proc Natl Acad Sci U.S.A. (1980) 77(12):7152–6. doi: 10.1073/pnas.77.12.7152
33. Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol (2003) 4(1):69–77. doi: 10.1038/ni875
34. Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol (2003) 4(1):63–8. doi: 10.1038/ni873
35. Platanias L. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol (2005) 5:375–86. doi: 10.1038/nri1604
36. Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet (2013) 5(2):164–71. doi: 10.1038/ng.2521
37. Nan Y, Nan G, Zhang YJ. Interferon induction by RNA viruses and antagonism by viral pathogens. Viruses (2014) 6(12):4999–5027. doi: 10.3390/v6124999
38. Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol (2014) 32:513–45. doi: 10.1146/annurev-immunol-032713-120231
39. Kotenko SV, Durbin JE. Contribution of type III interferons to antiviral immunity: location, location, location. J Biol Chem (2017) 292(18):7295–303. doi: 10.1074/jbc.R117.777102
40. Peterson ST, Kennedy EA, Brigleb PH, Taylor GM, Urbanek K, Bricker TL, et al. Disruption of type III interferon (IFN) genes ifnl2 and ifnl3 recapitulates loss of the type III IFN receptor in the mucosal antiviral response. J Virol (2019) 93(22):e01073–19. doi: 10.1128/JVI.01073-19
41. Broggi A, Granucci F, Zanoni I. Type III interferons: Balancing tissue tolerance and resistance to pathogen invasion. J Exp Med (2020) 217(1):e20190295. doi: 10.1084/jem.20190295
42. Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C. Antiviral activities of type I interferons to SARS CoV-2 infection. Antiviral Res (2020) 179:104811. doi: 10.1016/j.antiviral.2020.104811
43. Goel RR, Kotenko SV, Kaplan MJ. Interferon lambda in inflammation and autoimmune rheumatic diseases. Nat Rev Rheumatol (2021) 17(6):349–62. doi: 10.1038/s41584-021-00606-1
44. Alphonse N, Dickenson RE, Alrehaili A, Odendall C. Functions of IFNλs in anti-bacterial immunity at mucosal barriers. Front Immunol (2022) 13:857639. doi: 10.3389/fimmu.2022.857639
45. Chen SN, Gan Z, Hou J, Yang YC, Huang L, Huang B, et al. Identification and establishment of type IV interferon and the characterization of interferon-υ including its class II cytokine receptors IFN-υR1 and IL-10R2. Nat Commun (2022) 13(1):999. doi: 10.1038/s41467-022-28645-6
46. Antos D, Alcorn JF. IFNλ: balancing the light and dark side in pulmonary infection. mBio. (2023) 14(4):e0285022. doi: 10.1128/mbio.02850-22
47. Wang H, Hu H, Zhang K. Overview of interferon: characteristics, signaling and anti-cancer effect. Arch Biotechnol Biomed (2017) 1:001–16. doi: 10.29328/journal.hjb.1001001
48. Lazear HM, Schoggins JW, Diamond MS. Shared and distinct functions of type I and type III interferons. Immunity (2019) 50(4):907–23. doi: 10.1016/j.immuni.2019.03.025
49. Song K, Moon DB, Kim NY, Shin YK. Stability and activity of the hyperglycosylated human interferon-β R27T variant. Sci Rep (2020) 10:8412. doi: 10.1038/s41598-020-65495-y
50. Walter MR. The Role of structure in the biology of interferon signaling. Front Immunol (2020) 11:606489. doi: 10.3389/fimmu.2020.606489
51. Anjum FR, Rahman SU, Aslam MA, Qureshi AS. Antiviral potential and stability analysis of chicken interferon-α produced by Newcastle disease virus in chicken embryo fibroblast cells. Vet Med – Czech. (2021) 66(5):197–207. doi: 10.17221/106/2020-VETMED
52. Pisanelli G, Pagnini U, Iovane G, García-Sastre A. Type I and type II interferon antagonism strategies used by paramyxoviridae: previous and new discoveries, in comparison. Viruses. (2022) 14(5):1107. doi: 10.3390/v14051107
53. Gonzalez-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol (2012) 12(2):125–35. doi: 10.1038/nri3133
54. Mak TW, Saunders ME. Immunity to pathogens. Immune Response (2006), 641–94. doi: 10.1016/B978-012088451-3.50024-7
55. Lee AJ, Ashkar AA. The dual nature of type I and type II interferons. Front Immunol (2018) 9:2061. doi: 10.3389/fimmu.2018.02061
56. Schoggins WJ. IFN stimulated genes: what do they all do? Ann. Rev Virol (2019) 6:567–84. doi: 10.1146/annurev-virology-092818-015756
57. Ali S, Mann-Nüttel R, Schulze A, Richter L, Alferink J, Scheu S. Sources of type I interferons in infectious immunity: plasmacytoid dendritic cells not always in the driver's seat. Front Immunol (2019) 10:778. doi: 10.3389/fimmu.2019.00778
58. Chiale C, Greene TT, Zuniga EI. Interferon induction, evasion, and paradoxical roles during SARS CoV-2 infection. Immunol Rev (2022) 309(1):12–24. doi: 10.1111/imr.13113
59. Zhu Z, Yang X, Huang C, Liu L. The interferon-induced protein with tetratricopeptide repeats repress influenza virus infection by inhibiting viral RNA synthesis. Viruses. (2023) 15(7):1412. doi: 10.3390/v15071412
60. Chen YG, Hur S. Cellular origins of dsRNA, their recognition and consequences. Nat Rev Mol Cell Biol (2022) 23(4):286–301. doi: 10.1038/s41580-021-00430-1
61. Su CM, Du Y, Rowland RRR, Wang Q, Yoo D. Reprogramming viral immune evasion for a rational design of next-generation vaccines for RNA viruses. Front Immunol (2023) 14:1172000. doi: 10.3389/fimmu.2023.1172000
62. Halajian EA, LeBlanc EV, Gee K, Colpitts CC. Activation of TLR4 by viral glycoproteins: A double-edged sword? Front Microbiol (2022) 13:1007081. doi: 10.3389/fmicb.2022.1007081
63. Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol (2003) 21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126
64. Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol (2004) 4(7):499–511. doi: 10.1038/nri1391
65. Lester SN, Li K. Toll-like receptors in antiviral innate immunity. J Mol Biol (2014) 426(6):1246–64. doi: 10.1016/j.jmb.2013.11.024
66. Chasset F. Type I IFNs in systemic autoimmune diseases. Front Pharmacol (2021) 12:633821. doi: 10.3389/fphar.2021.633821
67. Saunders LR, Barber GN. The ds binding protein family. Critical roles, diverse cellular functions. FASEB (2003) 17(9):961–83. doi: 10.1096/fj.02-0958rev
68. Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, et al. 5'-triphosphate RNA is the ligand for RIG-I. Science (2006) 314(5801):994–7. doi: 10.1126/science.1132505
69. Schlee M. Master sensors of pathogenic RNA-RIG-I like receptors. Immunobiology (2013) 218(11):1322–35. doi: 10.1016/j.imbio.2013.06.007
70. Kell AM, Gale M Jr. RIG-I in RNA virus recognition. Virology (2015) 479-480:110–21. doi: 10.1016/j.virol.2015.02.017
71. Chan CP, Jin D-J. Cytoplasmic RNA sensors and their interplay with RNA-binding partners in innate antiviral response. RNA (2022) 28(4):449–77. doi: 10.1261/rna.079016.121
72. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science (2013) 339(6121):786–91. doi: 10.1126/science.1232458
73. Wu J, Sun L, Chen X, Du F, Shi H, Chen C, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science (2013) 339(6121):826–30. doi: 10.1126/science.1229963
74. Jia J, Fu J, Tang H. Activation and evasion of RLR signaling by DNA virus infection. Front Microbiol (2021) 12:804511. doi: 10.3389/fmicb.2021.804511
75. Mnich ME, van Dalen R, van Sorge NM. C-type lectin receptors in host defense against bacterial pathogens. Front Cell Infect Microbiol (2020) 10:309. doi: 10.3389/fcimb.2020.00309
76. Jang JH, Shin HW, Lee JM, Lee HW, Kim EC, Park SH. An overview of pathogen recognition receptors for innate immunity in dental pulp. Mediators Inflamm (2015) 2015:794143. doi: 10.1155/2015/794143
77. Caneparo V, Landolfo S, Gariglio M, De Andrea M. The absent in melanoma 2-like receptor IFN-inducible protein 16 as an inflammasome regulator in systemic lupus erythematosus: the dark side of sensing microbes. Front Immunol (2018) 9:1180. doi: 10.3389/fimmu.2018.01180
78. Fan Z, Chen R, Yin W, Xie X, Wang S, Hao C. Effects of AIM2 and IFI16 on infectious diseases and inflammation. Viral Immunol (2023) 36(7):438–48. doi: 10.1089/vim.2023.0044
79. Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signaling. Nat Rev Mol Cell Biol (2020) 21:501–21. doi: 10.1038/S41580-020-0244-x
80. Lin JY, Kuo RL, Huang HI. Activation of type I interferon antiviral response in human neural stem cells. Stem Cell Res Ther (2019) 10(1):387. doi: 10.1186/s13287-019-1521-5
81. Godecke N, Riedel J, Herrmann S, Behme S, Rand U, Kubsch T, et al. Synthetic rewiring and boosting type I interferon responses for visualization and counteracting viral infections. Nucleic Acids Res (2020) 48(20):11799–811. doi: 10.1093/nar/gkaa961
82. Gutierrez-Merino J, Isla B, Combes T, Martinez-Estrada F, Maluquer De Motes C. Beneficial bacteria activate type-I interferon production via the intracellular cytosolic sensors STING and MAVS. Gut Microbes (2020) 11(4):771–88. doi: 10.1080/19490976.2019.1707015
83. Rojas JM, Alejo A, Martín V, Sevilla N. Viral pathogen-induced mechanisms to antagonize mammalian interferon (IFN) signaling pathway. Cell Mol Life Sci (2021) 78(4):1423–44. doi: 10.1007/s00018-020-03671-z
84. Pestka S, Krause DC, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev (2004) 202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x
85. Krause CD, Lavnikova N, Xie J, Mei E, Mirochnitchenko OV, Jia Y, et al. Preassembly and ligand-induced restructuring of the chains of the IFN-gamma receptor complex: the roles of Jak kinases, Stat1 and the receptor chains. Cell Res (2006) 16(1):55–69. doi: 10.1038/sj.cr.7310008
86. Daza-Cajigal V, Albuquerque AS, Young DF, Ciancanelli MJ, Moulding D, Angulo I, et al. Partial human Janus kinase 1 deficiency predominantly impairs responses to interferon gamma and intracellular control of mycobacteria. Front Immunol (2022) 13:888427. doi: 10.3389/fimmu.2022.888427
87. Babon JJ, Lucet IS, Murphy JM, Nicola NA, Varghese LN. The molecular regulation of Janus kinase (JAK) activation. Biochem J (2014) 462(1):1–13. doi: 10.1042/BJ20140712
88. Stanifer ML, Pervolaraki K, Boulant S. Differential regulation of type I and type III interferon signaling. Int J Mol Sci (2019) 20(6):1445. doi: 10.3390/ijms20061445
89. de Weerd NA, Nguyen T. The interferons and their receptors–distribution and regulation. Immunol Cell Biol (2012) 90(5):483–91. doi: 10.1038/icb.2012.9
90. Park A, Iwasaki A. Type I and type III interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe (2020) 27(6):870–8. doi: 10.1016/j.chom.2020.05.008
91. Manivasagam S, Klein RS. Type III interferons: emerging roles in autoimmunity. Front Immunol (2021) 12:764062. doi: 10.3389/fimmu.2021.764062
92. Seif F, Khoshmirsafa M, Aazami H, Monireh Mohsenzadegan M, Sedighi G, Bahar M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun Signal (2017) 15:23. doi: 10.1186/s12964-017-0177-y
93. Mazewski C, Perez RE, Fish EN, Platanias LC. Type I interferon (IFN)-regulated activation of canonical and non-canonical signaling pathways. Front Immunol (2020) 11:606456. doi: 10.3389/fimmu.2020.606456
94. Su J. The discovery of type IV interferon system revolutionizes interferon family and opens up a new frontier in jawed vertebrate immune defense. Sci China Life Sci (2022) 65(11):2335–7. doi: 10.1007/s11427-022-2112-0
95. Paul A, Tang TH, Ng SK. Interferon regulatory factor 9 structure and regulation. Front Immunol (2018) 9:1831. doi: 10.3389/fimmu.2018.01831
96. Sekrecka A, Kluzek K, Sekrecki M, Boroujeni ME, Hassani S, Yamauchi S, et al. Time-dependent recruitment of GAF, ISGF3 and IRF1 complexes shapes IFNα and IFNγ-activated transcriptional responses and explains mechanistic and functional overlap. Cell Mol Life Sci (2023) 80(7):187. doi: 10.1007/s00018-023-04830-8
97. Low ZY, Zabidi NZ, Yip AJW, Puniyamurti A, Chow VTK, Lal SK. SARS CoV-2 Non-structural proteins and their roles in host immune evasion. Viruses (2022) 14(9):1991. doi: 10.3390/v14091991
98. Rashid F, Xie Z, Suleman M, Shah A, Khan S, Luo S. Roles and functions in SARS CoV-2 proteins in host immune evasion. Front Immunol (2022) 3:940756. doi: 10.3389/fimmu.2022.940756
99. Rehwinkel J, Gack MU. RIG-I like receptors. Nat Rev Immunol (2020) 20(9):537–51. doi: 10.1038/s41577-020-0283-3
100. Onomoto K, Onoguchi K, Yoneyama M. Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cell Mol Immunol (2021) 18(3):539–55. doi: 10.1038/s41423-020-00602-7
101. Uddin S, Yenush L, Sun XJ, Sweet ME, White MF, Platanias LC. Interferon-alpha engages the insulin receptor substrate-1 to associate with the phosphatidylinositol 3'-kinase. J Biol Chem (1995) 270(27):15938–41. doi: 10.1074/jbc.270.27.15938
102. Ivashkiv L, Donlin L. Regulation of type I interferon responses. Nat Rev Immunol (2014) 14:36–49. doi: 10.1038/nri3581
103. Watson A, Spalluto CM, McCrae C, Cellura D, Burke H, Cunoosamy D, et al. Dynamics of IFN-β Responses during respiratory viral infection. Insights for therapeutic strategies. Am J Respir Crit Care Med (2020) 201(1):83–94. doi: 10.1164/rccm.201901-0214OC
104. Stegelmeier AA, Darzianiazizi M, Hanada K, Sharif S, Wootton SK, Bridle BW, et al. Type I interferon-mediated regulation of antiviral capabilities of neutrophils. Int J Mol Sci (2021) 22(9):4726. doi: 10.3390/ijms22094726
105. de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol (2001) 69(6):912–20. doi: 10.1189/jlb.69.6.912
106. Chawla-Sarkar M, Lindner DJ, Liu YF, Williams BR, Sen GC, Silverman RH, et al. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis (2003) 8(3):237–49. doi: 10.1023/a:1023668705040
108. Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer. (2016) 16(3):131–44. doi: 10.1038/nrc.2016.14
109. Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature (2011) 472(7344):481–5. doi: 10.1038/nature09907
110. McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol (2015) 15(2):87–103. doi: 10.1038/nri3787
111. Yang E, Li MMH. All About the RNA: Interferon-stimulated genes that interfere with viral RNA processes. Front Immunol (2020) 11:605024. doi: 10.3389/fimmu.2020.605024
112. Ostrycharz E, Hukowska-Szematowicz B. Micro-players of great significance-host microRNA signature in viral infections in humans and animals. Int J Mol Sci (2022) 23(18):10536. doi: 10.3390/ijms231810536
113. Levine S. Effect of actinomycin D and puromycin dihydrochloride on action of interferon. Virology. (1964) 24(4):586–8. doi: 10.1016/0042-6822(64)90211-9
114. Wagner R. Inhibition of interferon biosynthesis by actinomycin D. Nature (1964) 204:49–51. doi: 10.1038/204049a0
115. Rauch I, Mȕller M, Decker T. The regulation of inflammation by IFNs and their STATS. JACSTAT (2013) 2(1):e23820. doi: 10.4161/jkst.23820
116. Sorgeloos F, Kreit M, Hermant P, Lardinois C, Michiels T. Antiviral type I and type III interferon responses in the central nervous system. Viruses (2013) 5(3):834–57. doi: 10.3390/v5030834
117. Mishra R, Kumar A, Ingle H, Kumar H. The interplay between viral-derived miRNAs and host immunity during infection. Front Immunol (2020) 10:3079. doi: 10.3389/fimmu.2019.03079
118. Singh M, Chazal M, Quarato P, Bourdon L, Malabat C, Vallet T, et al. A virus-derived microRNA targets immune response genes during SARS CoV-2 infection. EMBO Rep (2022) 23(2):e54341. doi: 10.15252/embr.202154341
119. Baum A, Garcia-Sastre A. Induction of Type I IFN by RNA viruses: cellular receptors and their substrates. Amino Acids (2010) 38(5):1283–89. doi: 10.1007/s00726-009-0374-0
120. Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in détente. Science (2006) 312(5775):879–82. doi: 10.1126/science.1125676
121. Dauber B, Wolff T. Activation of the antiviral kinase PKR and viral countermeasures. Viruses (2009) 1(3):523–44. doi: 10.3390/v1030523
122. Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res (2011) 31(1):41–7. doi: 10.1089/jir.2010.0107
123. Hasham K, Ahmed N, Zeshan B. Circulating microRNAs in oncogenic viral infections: potential diagnostic biomarkers. SN Appl Sci (2020) 2:442. doi: 10.1007/s42452-020-2251-0
124. Wang Z, Pan H, Jiang B. Type I IFN deficiency: an immunological characteristic of severe COVID-19 patients. Signal Transduct Target Ther (2020) 5(1):198. doi: 10.1038/s41392-020-00306-4
125. Fani M, Zandi M, Rezayi M, Khodadad N, Langari H, Amiri I. The Role of microRNAs in the viral infections. Curr Pharm Des (2018) 24(39):4659–67. doi: 10.2174/1381612825666190110161034
126. Fani M, Zandi M, Ebrahimi S, Soltani S, Abbasi S. The role of miRNAs in COVID-19 disease. Future Virol (2021) 16. doi: 10.2217/fvl-2020-0389
127. Lindqvist R, Mundt F, Gilthorpe JD, Wölfel S, Gekara NO, Kröger A, et al. Fast type I interferon response protects astrocytes from flavivirus infection and virus-induced cytopathic effects. J Neuroinflamm (2016) 13(1):277. doi: 10.1186/s12974-016-0748-7
128. Hwang M, Bergman C. IFNα/β signaling in astrocytes mediates protection against viral encephalomyelitis. J Virol (2018) 92:e01901–17. doi: 10.1128/JVI.01901-17
129. Teijaro JR. Type I interferons in viral control and immune regulation. Curr Opin Virol (2016) 16:31–40. doi: 10.1016/j.coviro.2016.01.001
130. Zhang SY, Boisson-Dupuis S, Chapgier A, Yang K, Bustamante J, Puel A, et al. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol Rev (2008) 226:29–40. doi: 10.1111/j.1600-065X.2008.00698.x
131. Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science (2001) 293(5531):834–8. doi: 10.1126/science.1062961
132. Schoggins WJ, Rice CM. ISG and their antiviral effector functions. Curr Opin Virol (2011) 1(6):519–25. doi: 10.1016/j.coviro.2011.10.008
133. Atri C, Guerfali FZ, Laouini D. Role of human macrophage polarization in inflammation during infectious diseases. Int J Mol Sci (2018) 19(6):1801. doi: 10.3390/ijms19061801
134. Tan HY, Wang N, Li S, Hong M, Wang X, Feng Y. The reactive oxygen species in macrophage polarization: reflecting its dual role in progression and treatment of human diseases. Oxid Med Cell Longev (2016) 2016:2795090. doi: 10.1155/2016/2795090
135. Weng HL, Cai WM, Liu RH. Animal experiment and clinical study of effect of gamma-interferon on hepatic fibrosis. World J Gastroenterol (2001) 7(1):42–8. doi: 10.3748/wjg.v7.i1.42
136. Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PloS Pathog (2008) 4(3):e1000017. doi: 10.1371/journal.ppat.1000017
137. Esposito MM, Turku S, Lehrfield L, Shoman A. The impact of human activities on zoonotic infection transmissions. Anim (Basel). (2023) 13(10):1646. doi: 10.3390/ani13101646
138. Hoag H. Study revives bird origin for 1918 flu pandemic. Nature (2014). doi: 10.1038/nature.2014.14723
139. Cook PW, Stark T, Jones J, Kondor R, Zanders N, Benfer J, et al. Detection and characterization of swine origin influenza A(H1N1) pandemic 2009 viruses in humans following zoonotic transmission. J Virol (2020) 95(2):e01066–20. doi: 10.1128/JVI.01066-20
140. Liu WJ, Xiao H, Dai L, Liu D, Chen J, Qi X, et al. (H7N9) virus: from low pathogenic to highly pathogenic. Front Med (2021) 15(4):507–27. doi: 10.1007/s11684-020-0814-5
141. Jani C, Patel K, Walker A, Singh H, Al Omari O, Crowley C, et al. Trends of HIV mortality between 2001 and 2018: an observational analysis. Trop Med Infect Dis (2021) 6(4):173. doi: 10.3390/tropicalmed6040173
142. Stasi C, Silvestri C, Voller F. Update on hepatitis C epidemiology: unaware and untreated infected population could be the key to elimination. SN Compr Clin Med (2020) 2(12):2808–15. doi: 10.1007/s42399-020-00588-3
143. Dudas G, Carvalho LM, Rambaut A, Bedford T. MERS-CoV spillover at the camel-human interface. Elife. (2018) 7:e31257. doi: 10.7554/eLife.31257
144. Hooper J. Contamination: the case of civets, companionship, COVID, and SARS. J Appl Anim Welf Sci (2022) 25(2):167–79. doi: 10.1080/10888705.2022.2028627
145. Gazal S, Sharma N, Gazal S, Tikoo M, Shikha D, Badroo GA, et al. Nipah and hendra viruses: deadly zoonotic paramyxoviruses with the potential to cause the next pandemic. Pathogens. (2022) 11(12):1419. doi: 10.3390/pathogens11121419
146. Lecollinet S, Pronost S, Coulpier M, Beck C, Gonzalez G, Leblond A, et al. Viral equine encephalitis, a growing threat to the horse population in europe? Viruses (2019) 12(1):23. doi: 10.3390/v12010023
147. Voskarides K. Animal-to-human viral transitions: is SARS coV-2 an evolutionarily successful one? J Mol Evol (2020) 88(5):421–3. doi: 10.1007/s00239-020-09947-z
148. Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol (2020) 17(5):533–5. doi: 10.1038/s41423-020-0402-2
149. Westover JB, Hickerson BT, Van Wettere AJ, Hurst BL, Kurz JP, Dagley A, et al. Vascular leak and hypercytokinemia associated with severe fever with thrombocytopenia syndrome virus infection in mice. Pathogens (2019) 8(4):158. doi: 10.3390/pathogens8040158
150. Hoagland DA, Møller R, Uhl SA, Oishi K, Frere J, Golynker I, et al. Leveraging the antiviral type I interferon system as a first line of defense against SARS CoV-2 pathogenicity. Immunity (2021) 54(3):557–570.e5. doi: 10.1016/j.immuni.2021.01.017
151. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I IFN activity and inflammatory responses in severe COVID-19 patients. Science (2020) 369:718–24. doi: 10.1126/science.abc6027
152. Wang W, Zhou Z, Xiao X, Tian Z, Dong X, Wang C, et al. SARS CoV-2 nsp12 attenuates type I interferon production by inhibiting IRF3 nuclear translocation. Cell Mol Immunol (2021) 18(4):945–53. doi: 10.1038/s41423-020-00619-y
153. Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science (2020) 370. doi: 10.1126/science.abd4585
154. Zhang Q, Bastard P, Bolze A, Jouanguy E, Zhang SY, COVID Human Genetic Effort, et al. Life-threatening COVID-19: defective interferons unleash excessive inflammation. Med. (2020) 1(1):14–20. doi: 10.1016/j.medj.2020.12.001
155. Channappanavar R, Fehr AR, Zheng J, Wohlford-Lenane C, Abrahante JE, Mack M, et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest. (2019) 129(9):3625–39. doi: 10.1172/JCI126363
156. Rebendenne A, Valadão ALC, Tauziet M, Maarifi G, Bonaventure B, McKellar J, et al. SARS CoV-2-triggers and MDA dependent IFN response which is unable to control replication in lung epithelial cells. J Virol (2021) 95:e02415–20. doi: 10.1128/JVI.02415-20
157. Karki R, Sharma BR, Tuladhar S, Williams EP, Zalduondo L, Samir P, et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS CoV-2 infection and cytokine shock syndromes. Cell (2021) 184(1):149–168.e17. doi: 10.1016/j.cell.2020.11.025
158. Schiuma G, Beltrami S, Bortolotti D, Rizzo S, Rizzo R. Innate immune response in SARS CoV-2 infection. Microorganisms (2022) 10(3):501. doi: 10.3390/microorganisms10030501
159. Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis (2020) 71(15):762–8. doi: 10.1093/cid/ciaa248
160. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol (2020) 20(6):363–74. doi: 10.1038/s41577-020-0311-8
161. Thorne LG, Reuschl AK, Zuliani-Alvarez L, Whelan MVX, Turner J, Noursadeghi M, et al. SARS CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. EMBO J (2021) 40(15):e107826. doi: 10.15252/embj.2021107826
162. Wolff D, Nee S, Hickey NS, Marschollek M. Risk factors for Covid-19 severity and fatality: a structured literature review. Infection (2021) 49(1):15–28. doi: 10.1007/s15010-020-01509-1
163. Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, et al. Imbalanced host response to SARS CoV-2 drives development of COVID-19. Cell (2020) 181(5):1036–1045.e9. doi: 10.1016/j.cell-2020.04.026
164. Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe (2020) 27(6):883–890.e2. doi: 10.1016/j.cho.2020.04.017
165. Shen WX, Luo RC, Wang JQ, Chen ZS. Features of cytokine storm identified by distinguishing clinical manifestations in COVID-19. Front Public Health (2021) 9:671788. doi: 10.3389/fpubh.2021.671788
166. Wall EM, Cao JX, Upton C. Subversion of cytokine networks by viruses. Int Rev Immunol (1998) 17(1-4):121–55. doi: 10.3109/08830189809084490
167. Kouwaki T, Nishimura T, Wang G, Oshiumi H. RIG-I-like receptor-mediated recognition of viral genomic RNA of severe acute respiratory syndrome coronavirus-2 and viral escape from the host innate immune responses. Front Immunol (2021) 12:700926. doi: 10.3389/fimmu.2021.700926
168. Sampaio NG, Chauveau L, Hertzog J, Bridgeman A, Fowler G, Moonen JP, et al. The RNA sensor MDA5 detects SARS CoV-2 infection. Sci Rep (2021) 11(1):13638. doi: 10.1038/s41598-021-92940-3
169. Yin X, Riva L, Pu Y, Martin-Sancho L, Kanamune J, Yamamoto Y, et al. MDA5 governs the innate immune response to SARS CoV-2 in lung epithelial cells. Cell Rep (2021) 34(2):108628. doi: 10.1016/j.celrep.2020.108628
170. Ulrich D, Gray J. Rotaviruses. In: Zuckerman AJ, Banatvala JE, Pattison JR, Griffith PD, Schoub BD, editors. Principles and Practice of Clinical Virology, fifth Edition. (England: John Wiley & Sons, Ltd) (2004). 912 p.
171. Angel J, Franco MA. Rotaviruses, in Desk Encyclopedia of Human and Medical Virology. Mahy BWJ, Regenmortel MHV, editors. (Cambridge, Massachusetts, USA: Academic Press) (2008).
172. Akira S, Uematsu, Takeuchi O. Pathogen recognition and innate immunity. Cell (2006) 124(4):783–801. doi: 10.1016/jcell.2006.02.015
173. Nelemans T, Kikkert M. Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses. (2019) 11(10):961. doi: 10.3390/v11100961
174. Dzananovic E, McKenna SA, Patel TR. Viral proteins targeting host protein kinase R to evade an innate immune response: a mini review. Biotechnol Genet Eng Rev (2018) 34(1):33–59. doi: 10.1080/02648725.2018.1467151
175. Cesaro T, Michiels T. Inhibition of PKR by viruses. Front Microbiol (2021) 12. doi: 10.3389/fmicb.2021.757238
176. Seet BT, Johnston JB, Brunetti CR, Barrett JW, Everett H, Cameron C, et al. Poxviruses and immune evasion. Annu Rev Immunol (2003) 21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049
177. Brussow H. Immunology of COVID-19. Environ Microbiol (2020) 22:4895–908. doi: 10.1111/1462-2920.15302
178. Gorbunova V, Seluanov A, Kennedy BK. The world goes bats: living longer and tolerating viruses. Cell Metab (2020) 32(1):31–43. doi: 10.1016/j.cmet.2020.06.013
179. Sui L, Zhao Y, Wang W, Wu P, Wang Z, Yu Y, et al. SARS CoV-2 membrane protein inhibits type I interferon production through ubiquitin-mediated degradation of TBK1. Front Immunol (2021) 12:662989. doi: 10.3389/fimmu.2021.662989
180. Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol (2008) 82(9):4471–9. doi: 10.1128/JVI.02472-07
181. Kamitani W, Huang C, Narayanan K, Lokugamage KG. Makino S.A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol (2009) 16:1134–40. doi: 10.1038/nsmb.1680
182. Han L, Zhuang MW, Deng J, Zheng Y, Zhang J, Nan ML, et al. SARS CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways. J Med Virol (2021) 93(9):5376–89. doi: 10.1002/jmv.27050
183. Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, et al. Evasion of type I interferon by SARS CoV-2. Cell Rep (2020) 33(1):108234. doi: 10.1016/j.celrep.2020.108234
184. Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, et al. Activation and evasion of type I IFN responses by SARS CoV-2. Nat Commun (2020) 11:3810. doi: 10.1038/s41467-020-17665-9
185. Chow KT, Gale M Jr, Loo YM. RIG-I and other RNA sensors in antiviral immunity. Annu Rev Immunol (2018) 36:667–94. doi: 10.1146/annurev-immunol-042617-053309
186. Wadman M. Public needs to prep for vaccine side effects. Science (2020) 370(6520):1022. doi: 10.1126/science.370.6520.1022
187. Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS CoV-2 in clinical samples. Lancet Infect Dis (2020) 20(4):411–2. doi: 10.1016/S1473-3099(20)30113-4
188. Yu Y, Luo D, Chen X, Huang Z, Wang M, Xiao S. Medication adherence to antiretroviral therapy among newly treated people living with HIV. BMC Public Health (2018) 18(1):825. doi: 10.1186/s12889-018-5731-z
189. Hasselbalch HC, Skov V, Kjær L, Ellervik C, Poulsen A, Poulsen TD, et al. COVID-19 as a mediator of interferon deficiency and hyperinflammation: Rationale for the use of JAK1/2 inhibitors in combination with interferon. Cytokine Growth Factor Rev (2021) 60:28–45. doi: 10.1016/j.cytogfr.2021.03.006
190. Rarani FZ, Rashidi B, Jafari Najaf Abadi MH, Hamblin MR, Reza Hashemian SM, Mirzaei H. Cytokines and microRNAs in SARS-CoV-2: What do we know? Mol Ther Nucleic Acids (2022) 29:219–42. doi: 10.1016/j.omtn.2022.06.017
191. Myasnikov AL, Berns SA, Talyzin PA, Ershov FI. [Interferon gamma in the treatment of patients with moderate COVID-19]. Voprosy Virusologii (2021) 66(1):47–54. doi: 10.36233/0507-4088-24
192. van Laarhoven A, Kurver L, Overheul GJ, Kooistra EJ, Abdo WF, van Crevel R, et al. Interferon gamma immunotherapy in five critically ill COVID-19 patients with impaired cellular immunity: A case series. Med. (2021) 2(10):1163–1170.e2. doi: 10.1016/j.medj.2021.09.003
193. Roquilly A, Francois B, Huet O, Launey Y, Lasocki S, Weiss E, et al. Interferon gamma-1b for the prevention of hospital-acquired pneumonia in critically ill patients: a phase 2, placebo-controlled randomized clinical trial. Intensive Care Med (2023) 49(5):530–44. doi: 10.1007/s00134-023-07065-0
194. Feld JJ, Kandel C, Biondi MJ, Kozak RA, Zahoor MA, Lemieux C, et al. Peginterferon lambda for the treatment of outpatients with COVID-19: a phase 2, placebo-controlled randomised trial. Lancet Respir Med (2021) 9(5):498–510. doi: 10.1016/S2213-2600(20)30566-X
195. Jagannathan P, Andrews JR, Bonilla H, Hedlin H, Jacobson KB, Balasubramanian V, et al. Peginterferon Lambda-1a for treatment of outpatients with uncomplicated COVID-19: a randomized placebo-controlled trial. Nat Commun (2021) 2(1):1967. doi: 10.1038/s41467-021-22177-1
Keywords: interferons, receptors, signal transduction pathways, SARS-CoV-2, therapeutic role of IFNs
Citation: Mihaescu G, Chifiriuc MC, Filip R, Bleotu C, Ditu LM, Constantin M, Cristian R-E, Grigore R, Bertesteanu SV, Bertesteanu G and Vrancianu CO (2024) Role of interferons in the antiviral battle: from virus-host crosstalk to prophylactic and therapeutic potential in SARS-CoV-2 infection. Front. Immunol. 14:1273604. doi: 10.3389/fimmu.2023.1273604
Received: 06 August 2023; Accepted: 29 December 2023;
Published: 15 January 2024.
Edited by:
Tanushree Dangi, Northwestern University, United StatesReviewed by:
Yongkui Li, Jinan University, ChinaDhiraj Kumar Singh, Texas Biomedical Research Institute, United States
Copyright © 2024 Mihaescu, Chifiriuc, Filip, Bleotu, Ditu, Constantin, Cristian, Grigore, Bertesteanu, Bertesteanu and Vrancianu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mariana Carmen Chifiriuc, carmen.chifiriuc@bio.unibuc.ro
 Grigore Mihaescu1,2
Grigore Mihaescu1,2 Mariana Carmen Chifiriuc
Mariana Carmen Chifiriuc Roxana Filip
Roxana Filip Coralia Bleotu
Coralia Bleotu Lia Mara Ditu
Lia Mara Ditu Marian Constantin
Marian Constantin Roxana-Elena Cristian
Roxana-Elena Cristian Corneliu Ovidiu Vrancianu
Corneliu Ovidiu Vrancianu