- Department of Oncology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
N6-methyladenosine (m6A) methylation is one of the most common modifications of RNA in eukaryotic cells, and is mainly regulated by m6A methyltransferases (writers), m6A demethylases (erasers), and m6A binding proteins (readers). Recently, accumulating evidence has shown that m6A methylation plays crucial roles in the regulation of the tumor immune microenvironment, greatly impacting the initiation, progression, and metastasis processes of various cancers. In this review we first briefly summarizes the m6A-related concepts and detection methods, and then describes in detail the associations of m6A methylation modification with various tumor immune components especially immune cells (e.g., regulatory T cells, dendritic cells, macrophages, and myeloid-derived suppressor cells) in a variety of cancers. We discuss the relationship between m6A methylation and cancer occurrence and development with the involvement of tumor immunity highlighted, suggesting novel markers and potential targets for molecular pathological diagnosis and immunotherapy of various cancers.
Introduction
N6-methyladenosine (m6A) methylation is one of the most abundant RNA modifications in eukaryotic cells (1, 2). It involves three kinds of vital regulatory proteins, namely writers, erasers, and readers (2), which can respectively add, remove, and preferentially bind to the m6A modification sites and modulate the fate of RNA (1). m6A modification is a key regulator of diverse RNA biology processes (Figure 1), including RNA processing, translation, stabilization, splicing, and degradation (3, 4). Recently, accumulating evidence has revealed the potential links between m6A modification and cancer immunology (5, 6). m6A modification plays vital roles in diverse tumor immunity processes among a variety of cancers, affecting the development, proliferation, growth, invasion, and metastasis of cancers (5). The tumor microenvironment (TME) is the environment around tumor cells, including the surrounding blood vessels, immune cells, fibroblasts, molecules, the extracellular matrix, and other stromal components. In this review, we discuss the characteristics of m6A modulators and the immunomodulatory function of m6A methylation in the tumor immune microenvironment (TIME), which refers to the immune and immune-associated components of the TME, and which is a complex interactive network consisting of various immune cells, cytokines, and fibroblasts that plays important roles in tumor initiation, progression, metastasis, and treatment response (7, 8). We focus on the associations of m6A modification in the TIME with cancer immunity and immunotherapy.
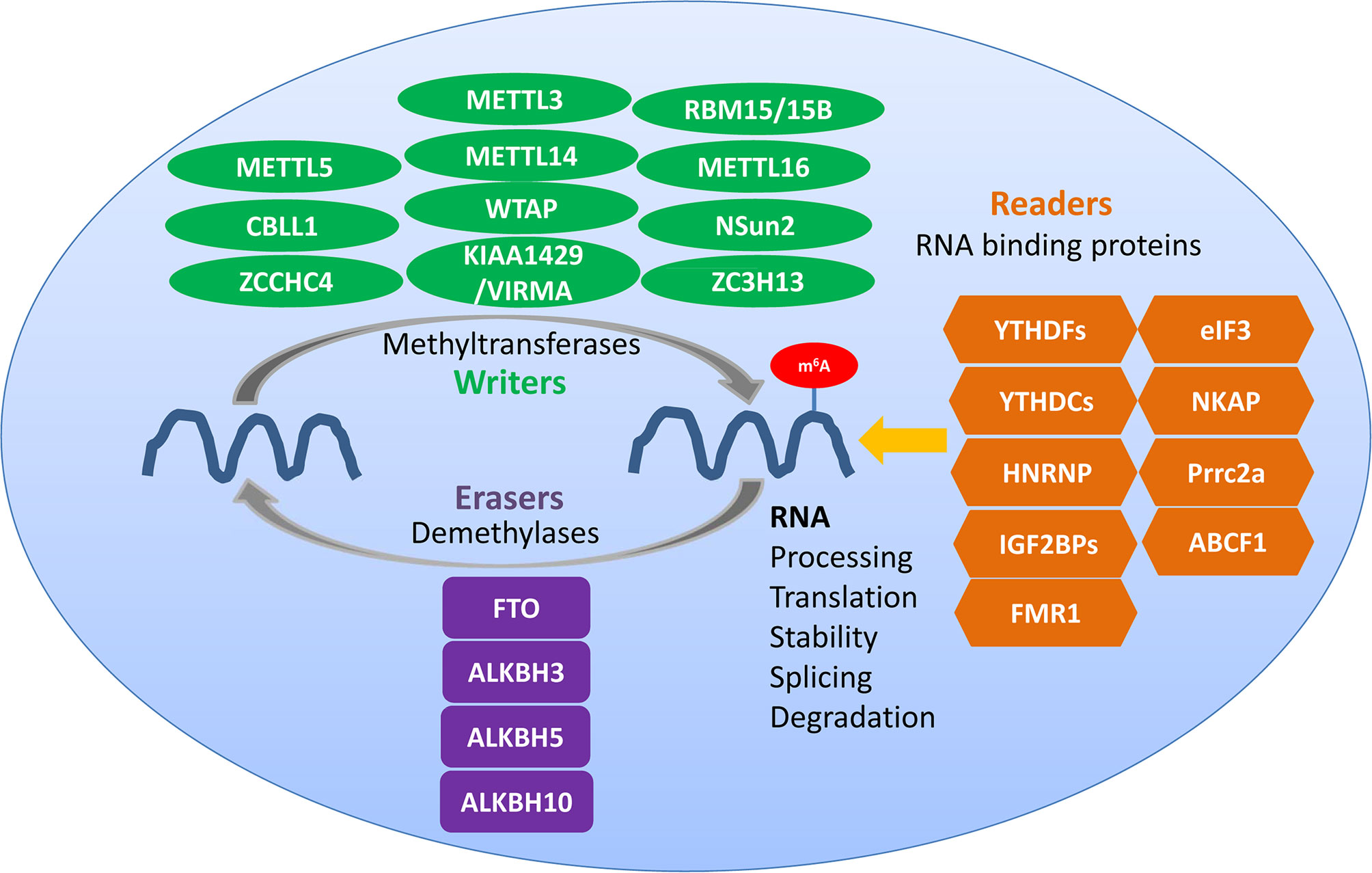
Figure 1 Regulators of m6A methylation within immune cells. Writers, erasers, and readers play different roles in the dynamic m6A modification of RNA. Common writers include METTL3, METTL14, and WTAP, common erasers include FTO and ALKBH5, and common readers include YTHDF1/2/3, YTHDC1/2, HNRNP, and IGF2BPs.
Regulation of m6A Methylation by m6A Writers, Erasers, and Readers
m6A Writers
m6A writers are generally considered to be composed of m6A methylases, majorly including methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), Wilms’ tumor 1-associating protein (WTAP), RNA binding motif protein 15 (RBM15) and its paralog RBM15B, which form the methyltransferase complex (MTC) (9). m6A writers are responsible for writing methylation information into RNA (6, 10). METTL3, a 70-kDa protein of the first identified m6A methyltransferases in eukaryotes, is a key enzymatic component of the MTC (6, 11); it can combine with S-adenosyl methionine (SAM) and transfer a methyl group to RNA. METTL4 is responsible for the recognition of substrates and functions as an allosteric activator that also binds to the target RNA (10–12). METTL14 serves as the RNA-binding platform, promoting the translation of related genes and enhancing the complex integrity (13, 14). METTL3 can form a heterodimer complex with the homologous protein METTL14. The METTL3-METTL14 dimer complex induces m6A deposition in transcripts on nuclear RNA (10). RBM15/15B interacts with METTL3 in a WTAP-dependent manner to help recruit the complexes to methylate-specific sites (15). Other m6A methyltransferases such as methyltransferase-like 16 (METTL16), zinc finger CCCH domain-containing protein 13 (ZC3H13), KIAA1429 [also known as vir-like m6A methyltransferase associated (VIRMA)], and NOP2/Sun domain family, member 2 (NSun2) are also essential for the formation of the MTC (16–21).
m6A Erasers
Obesity-associated protein FTO and alkB homolog 5 (ALKBH5) are m6A demethylases which are also called erasers and which ensure that m6A modification is a dynamic and reversible process (22). The RNA m6A modification can be removed by the demethylases FTO and ALKBH5. FTO was the first protein discovered to catalyze m6A demethylation, and knocking down the expression of FTO can increase the m6A methylation level of RNA. In contrast, when FTO is overexpressed, the m6A level of intracellular RNA is suppressed (23). FTO is located in nuclear speckles, where the m6A MTC also locates, and can reverse the m6A modification of RNA. ALKBH5 was the second m6A demethylase identified that could oxidatively reverse m6A modifications (22, 24). It is a Fe2+- and α-ketoglutarate-dependent non-heme oxygenase that can oxidize the N-methyl group at the m6A methylation site to hydroxymethyl group. ALKBH5 is also mainly located in nuclear speckles, and depends on its demethylase activity to affect the transport of RNA out of the nucleus; it then further modulates nuclear RNA metabolism and gene expression (22–25).
m6A Readers
The RNA-binding proteins that bind to m6A modification sites are called m6A readers, which include the YTH domain family (YTHDF1/2/3 and YTHDC1/2), heterogeneous nuclear ribonucleoproteins (HNRNPs; hnRNPC, hnRNPG, and hnRNPA2B1), and insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1-3); they can specifically bind to the m6A methylation sites affecting RNA metabolism, and are responsible for reversing or eliminating the RNA modification (23, 26–31). YTHDF1 interacts with translation initiation factors to promote translation and to reduce the binding of ribosomes to m6A-modified RNA, which promotes the degradation of RNA. YTHDF2 accelerates the decay of m6A-methylated RNA, and YTHDF3 can promote the translation promoted by YTHDF1 and regulate the YTHDF2-mediated RNA-decay-promotion (13, 29). The recognition of the ribonucleoprotein HNRNPC/G and its binding to the m6A modification sites are also indirect, which are mediated by the m6A switch mechanism and which participate in the processing and maturation of targeted RNA (13). The RNA-binding protein HNRNPA2B1 can bind to the nuclear m6A-modified RNA, allowing genes to be spliced (13, 22, 26). IGF2BP1-3 can recognize and bind to m6A modification sites, which increases the stability of target RNA and which promotes its translation (10).
Techniques for Detecting m6A Modifications
As early as in the 1970s, m6A methylation was identified to modify the mRNA and long non-coding RNA (lncRNA) in eukaryotes (11). However, limited by technical means, the detection especially the quantification of m6A and the identification of m6A at the single-base level had been progressing slowly (11, 32). The revitalization of researches related to m6A modification benefits from the emergence of effective analytical methods. The rapid development of next-generation sequencing (NGS) technologies and the improvement of liquid chromatography sensitivity provide a reliable basis for studying the influence of m6A RNA methylation on RNA structure (32–35).
An emerging method called methylated RNA immunoprecipitation sequencing (MeRIP-seq) or m6A-seq for identifying m6A modification sites on mammalian RNA emerged in 2012, and has recently received widespread attention (34–37). This new method is based on the high specificity of antibodies against m6A, and its combination with high-throughput sequencing makes it possible to describe the specific map of m6A modification in the mammalian transcriptome. The first step of MeRIP-seq is to fragment the RNA, followed by the use of immuno-magnetic beads with m6A antibody to enrich the m6A-methylated RNA fragments and the purification of the enriched RNA fragments to construct a high-throughput sequencing library by performing on-machine sequencing. In addition, a common transcriptome library needs to be constructed separately as a control. Finally, the two sequencing libraries are put together for bioinformatics analyses, and the region with a higher degree of m6A methylation is obtained, which is also called m6A peak (36, 37). The advantage of this method is that it is convenient, fast, and of low cost, and that it can enable a qualitative analysis of the RNA regions that are hyper-methylated. However, MeRIP-seq can only identify the m6A sites within RNA fragments of 100-200 nucleotides, and cannot achieve single-base resolution (34). To overcome the low resolution issue, a novel method called m6A individual nucleotide resolution crosslinking immunoprecipitation (miCLIP) has marked a major advancement in the field of m6A sequencing. This method enhances m6A-seq by UV-induced crosslinking of antibodies with immuno-precipitated RNA fragments (35, 38). Other approaches with higher resolutions include site-specific cleavage and radioactive-labelling followed by ligation-assisted extraction and thin-layer chromatography (SCARLET) and photo-crosslinking-assisted m6A-sequencing (PA-m6A-Seq) (38). Furthermore, it is currently possible to use the CRISPR-based genetic engineering modification methods to directly change any modification site in many organisms and to help develop m6A RNA methylation into a research method for more extensive investigations (34).
New breakthroughs have been made in the methods for detecting the overall m6A methylation level of cells, including the m6A dot-blot and high-performance liquid chromatography-mass spectrometry (HPLC-MS/MS) method to detect the overall m6A level of RNA, which can be used to generate important quantitative information on the presence and abundance of m6A modifiers (34).
m6A Modification as a Novel Regulator of the Tumor Immune Microenvironment
Recently, a growing number of studies have emerged mainly focusing on the mechanisms of strengthening anticancer immunity activation. The immune system is divided into innate and adaptive immunity. m6A plays critical roles in both the innate and adaptive immune responses and in tumor immunology, which provides important hints for developing various types of antitumor immunotherapies (22, 31). m6A also plays a vital role in the complex regulatory network within the TIME, and subsequently affects tumor occurrence, progression, metastasis, and treatment response (39–42). Most of the anticancer immune regulations rely on overcoming the continuous suppression of the adaptive immune response within the TME (43). Immune suppression is a typical feature of the TME, which involves the dysfunction of antigen presenting cells (APCs), recruitment or induction of large numbers of suppressive immune cells, such as CD4+ regulatory T cells (Tregs), dendritic cells (DCs), tumor-associated macrophages (TAMs), and myeloid cell-derived suppressor cells (MDSCs), and secretion of various cytokines (Figure 2) (44, 45).
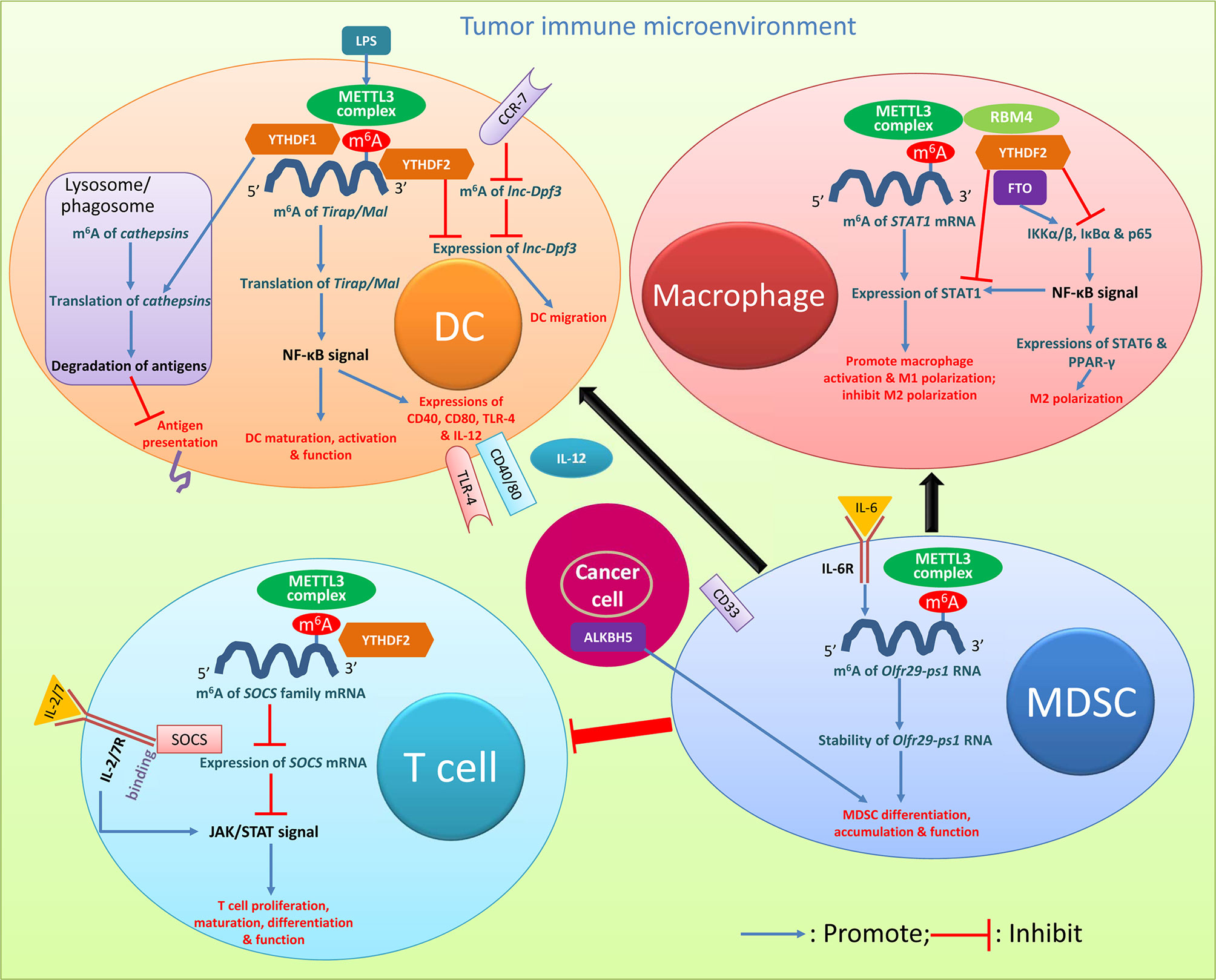
Figure 2 Roles of m6A modification in cancer immune regulation. Key m6A regulators and relevant pathways and molecules with biology activities clarified in various immune cells are presented. The descriptions are detailed in the relevant texts with citation of this figure. DC, dendritic cell; MDSC, myeloid-derived suppressor cell; LPS, lipopolysaccharide.
T and B Cells
T cells regulate the entire adaptive immune response. m6A is selectively regulated in tumor infiltrating T cells, and can be an important target in antitumor immunotherapies (44). Regulatory T cells (Tregs) are an important type of T cells which are involved in suppressing inflammation and producing immunosuppression (39). They are a key subset of effector T cells with strong immunosuppressive effects in the TME, and m6A-dependent immune function regulation was found in Tregs (44). The family of cytokine signal transduction (SOCS) proteins, mainly including SOCS1-3 and CIS, are inhibitory proteins involved in the transduction of the JAK-STAT signaling pathway, and play a vital role in inhibiting T cell proliferation and differentiation (39, 40, 43–48). The SOCS family genes are regulated by m6A methylation (44, 49); the modification was induced by the METTL3 complex at the 3’-end of the SOCS2 transcript, which could be recognized by YTHDF2 (48). Considering the importance of SOCS2 in Tregs in their noteworthy suppressive potential of the differentiation and function of CD4+ T cells and tumor-killing CD8+ T cells in the TME, the roles of m6A methylation of SOCS2 mRNA in T cells in immune response disorders during tumorigenesis should be further explored. In the process of inducing naive T cell development through IL-7 stimulation, the SOCS gene controls the IL-7 signaling (46). The SOCS family could act as a mediator binding to IL-7 receptor, which prevents STAT5 activation and inhibits downstream signals involved in T cell maturation and differentiation; T cells are highly responsive to IL-7 signals with downregulation of SOCS gene expressions, while upregulation of SOCS gene expressions suppresses the IL-7-dependent function of T cells (46). If naive CD4+ T cells are co-cultivated with METTL3-knockout Treg cells, naive T cells will have a greater ability to proliferate due to the complete lack of Treg inhibitory function (11, 50). In CD4+ T cells, reduced m6A modification can enhance the stability of the SOCS gene mRNA, thereby preventing signal transduction in the IL-2/STAT5 signaling pathway (11, 48, 50). The downregulation of METTL3 makes T cells stay in the naive T cell stage for longer time with reduced METTL3-mediated m6A methylation targeting the IL-7/STAT5/SOCS pathway (47). Together, m6A modification specifically targets the same class of genes encoding components of essential signaling pathways in different T cell subtypes; m6A methylation of SOCS mRNA with the involvement of m6A modulators including METTL3 and YTHDF2 decreases the stability and expression of SOCS mRNA, and reduces the inhibition of SOCS on the JAK/STAT signal, which promotes T cell proliferation, maturation differentiation, and function (Figure 2).
Downregulation of the m6A writer METTL14 specifically in B cells could result in severe defects in B cell development, with inhibition of IL-7-induced pro-B cell proliferation and blocking of large-to-small pre-B cell transition (51). More studies are needed to further reveal the regulatory roles of m6A modification in B cells.
Dendritic Cells (DCs)
Abundant abnormalities of m6A mRNA modifications were found in DCs in cancers. DCs are specialized APCs, which are responsible for the processing and presentation of antigens and the activation of T cell immune responses (11, 39). The regulation of their functions is closely related to the overall balance of immune responses. Exploring the regulatory mechanism of DC function activation is of great significance for in-depth understanding of the inflammatory and immune processes within the TME and for finding potential therapeutic targets for cancers with the involvement of abnormal DC activation (48, 52, 53). DCs have different stages of maturity: Immature DCs induce immune tolerance, mature DCs activate and stimulate immune response, and regulatory DCs downregulate immune response by suppressing T cell response (53). m6A methylation mediated by the methyltransferase METTL3 promotes the activation and function of DCs (54). Lipopolysaccharide can induce high expression levels of METTL3 in DCs (55). The specific consumption of METTL3 in DCs leads to impaired phenotype and functional maturation of DCs, and reduces the expression of costimulatory molecules CD40, CD80, and cytokine IL-12 involved in DC maturation. In vitro and in vivo studies have confirmed that after silencing METTL3 the ability of DCs to stimulate T cell responses is reduced. The METTL3-mediated m6A methylation modification of the CD40, CD80, and TLR4 signal transduction junction Tirap (also known as Mal) transcript enhances the translation in DCs to stimulate T cell activation and enhances the TLR4/NF-κB signal transduction to promote cytokine production; these confirm the new role of METTL3-mediated m6A methylation in promoting DC maturation (54). Furthermore, YTHDF1-knockdown mice had stronger response to tumor antigen-specific CD8+ T cells than wild-type mice; knockdown of YTHDF1 in classic DC cells enhanced the cross-presentation of tumor antigens in vivo and the cross-activation of CD8+ T cells (30, 40). The transcripts of multiple DC lysosomal cathepsins all have m6A modification, and YTHDF1 can promote the translation of lysosomal cathepsin in DCs by combining these transcripts (30, 56). The depletion of YTHDF1 in DCs attenuated the translation of genes related to the phagosome and lysosomal pathways which are a member of the cathepsin family (56). The enzymatic degradation of the proteins in phagosomes after DCs ingestion could destroy antigens to limit cross-presentation of antigens (57–59). The antigen cross-presentation of tumor-infiltrating DCs in YTHDF1-deficient melanoma or colon cancers can induce stronger anticancer immune response, and both in vitro and in vivo, mature YTHDF1-deficient DCs can induce stronger T cell activation than wild-type cells (57). YTHDF2 is also considered as a potential suppressor of tumor immunity (44). The activation of antitumor T cells and the initiation of anticancer immunity depend on the migration of APCs, especially the migration of DCs to lymph nodes (44, 60). CC-chemokine receptor 7 (CCR7) can stimulate the rapid but transient migration of DCs to draining lymph nodes, via upregulating the expression of the lncRNA lnc-Dpf3 by removing the m6A modification to prevent RNA degradation (61). DC-specific lnc-Dpf3 promotes CCR7-mediated DC migration, leading to excessive adaptive immune responses and inflammatory damage. The m6A-dependent manner regulates the dynamic expression of lnc-Dpf3 in DCs, and YTHDF2 reduces the expression level of lnc-Dpf3 in resting mature DCs (61). Together, in DCs m6A methylation of Tirap/Mal mRNA induced majorly by METTL3 strengthened the NF-κB signal, which promotes the expressions of several costimulatory molecules and enhances the maturation, activation, and function of DCs; the m6A regulator YTHDF1 promotes the translation of cathepsins in lysosomes/phagosomes, which enhances the degradation of antigens thus preventing antigen presentation and the subsequent activation of effector immune cells; YTHDF2 inhibits the expression of lnc-Dpf3, and suppresses the CCR-7-induced DC migration (Figure 2). The activation of DCs through m6A modification has vital roles in promoting the subsequent immune cell activation and function and in the migration of immune cells, thus importantly impacting the initiation and progression of cancers. The breakthrough of researches on the m6A epi-transcriptome accelerates the understanding of m6A-dependent DC development and activation, serving as prerequisite for future translational exploitation of m6A-based immunotherapy. Checkpoint blockade inducing YTHDF1 and/or YTHDF1 depletion in DCs may be a potential immunotherapy strategy (56).
Macrophages
Macrophages are immune cells derived from the hematopoietic system; they provide important innate immune defenses and maintain tissue-specific functions through regulation of the internal environment within organs (62, 63). For innate immunity, macrophages participate in the regulation of tissue homeostasis and resist viral infection and inflammation (48). Regarding the roles in inflammation and the TME, macrophages can be mainly polarized into the classically activated macrophages (M1 type) with antitumor function and the alternatively activated macrophages (M2 type), the latter of which can inhibit inflammation, promote angiogenesis and tissue repair, and also participate in tumor metastasis (48, 62–67). Upregulation of METTL3 activity greatly promotes the polarization of M1 macrophages, but it has an inhibitory effect on the polarization of M2 macrophages (68). METTL3 directly methylates the mRNA encoding signal transducer and activator of transcription 1 (STAT1), which is the main transcription factor that regulates the polarization into M1 macrophages (57). METTL3-mediated methylation of STAT1 mRNA significantly increases the mRNA stability and subsequently increases the expression of STAT1. METTL3 may serve as an anti-inflammatory target which drives the polarization of M1 macrophages by directly methylating STAT1 mRNA (68). Silencing the demethylase FTO significantly inhibits the polarization of M1 and M2 macrophages; FTO knockdown reduces the phosphorylation levels of IKKα/β, IκBα, and p65 in the NF-κB signaling pathway, which in turn leads to the downregulation of STAT1 expression in M1-type macrophages, and of STAT6 and peroxisome proliferation-activated receptor-γ (PPAR-γ) in M2-type macrophages. The actinomycin D experiment showed that silencing FTO could increase the instability of STAT1 and PPAR-γ mRNAs, thereby inhibiting transcription (64). Moreover, when the m6A reading protein YTHDF2 is silenced, the mRNA stability and expression of STAT1 and PPAR-γ increase. Silencing FTO can inhibit the NF-κB signaling pathway and reduce the stability of STAT1 and PPAR-γ mRNA through the participation of the YTHDF2 protein, thereby hindering the activation of macrophages (64, 65). FTO contributed to both M1 and M2 macrophage activation. RNA-binding motif 4 (RBM4) interacts with YTHDF2 and can be a possible inhibitor of M1 macrophage polarization via the degradation of m6A-modified STAT1 mRNA (69, 70). Together, in macrophages m6A methylation of STAT1 mRNA induced by METTL3 increases the expression of STAT1 and promotes macrophage activation and polarization into the M1 type; the m6A eraser FTO can strengthen the NF-κB signal, and promote both the expression of STAT1 for macrophage polarization into M1 type and the expressions of STAT6 and PPAR-γ for polarization into M2 type; YTHDF2 and RBM4 appear to have effects opposite to FTO (Figure 2). m6A modification majorly impacts the polarization of macrophages thus regulating cancer biology behaviors. These findings may open up new ways to study macrophage polarization and the underlying molecular mechanisms of its involvement in cancers.
Myeloid-Derived Suppressor Cells (MDSCs)
MDSCs are a group of heterogeneous myeloid cells generally with positive expression of CD33 and with potent immune-inhibitory activity; they have been identified as potential precursors of DCs, macrophages, and granulocytes (71, 72). In the TME MDSCs can suppress immune cells and protect tumors (73). The increase in METTL3 levels in CD33+ MDSCs in the TME is associated with poor prognosis (74). Knocking down METTL3 in CD33+ cells could attenuate MDSC or tumor-related MDSC differentiation in vitro (72). In MDSCs the lncRNA pseudogene Olfr29-ps1 was upregulated by the pro-inflammatory cytokine IL-6; the function of Olfr29-ps1 depended on IL-6-mediated m6A modification, and the lncRNA promoted the differentiation and immunosuppressive function of mononuclear MDSCs both in vitro and in vivo (74). ALKBH5 knockout in tumor cells enhanced the efficacy of immunotherapy, and ALKBH5 could regulate target gene expression and splicing, resulting in changes in metabolite contents and the accumulation of MDSCs (75). Together, METTL3-induced m6A methylation of Olfr29-ps1 which can be stimulated by IL-6 increases the stability of Olfr29-ps1, and promotes MDSC differentiation and function (Figure 2). The inhibition of MDSCs through the modification of m6A methylation may represent a promising anticancer treatment strategy.
Associations of m6A RNA Methylation With Tumor Immunity in Various Cancers
RNA methylation plays an important role in tumor genesis and development. Aberrant RNA methylation has been linked to various human cancers. The expression disorder of m6A RNA methylation regulators is closely related to a variety of cancers (48, 76, 77). RNA methylation affects tumor biology by regulating the relevant components of the immune system. It has been found that m6A RNA methylation has a variety of biology-regulatory functions during the occurrence and development of cancers via modulating tumor immunity (75, 78, 79). The TIME is often characterized by the infiltration of various immunosuppressive cell types, most notably MDSCs and Tregs, and a lack of antitumor immune activity (8, 80). In this section, we summarize the roles of m6A modulators in the TIME and immunotherapy within a variety of cancers, and organize the tumor types according to the anatomic systems which the involved organs belong to (nervous system, digestive system, respiratory system, urinary system, reproductive system, hematologic system, and others).
Nervous System Cancers
Glioblastoma and Glioma
In recent years, a large number of studies have proved that the TIME plays a vital role in cancer progression and anticancer therapeutic effects in glioblastoma and glioma (81, 82). Immune cells can penetrate into the brain and form an immune microenvironment. m6A regulatory factors are involved in multiple biological processes of tumor progression (50, 83–85). Therefore, clarifying the relationship between m6A regulatory factors and TME-infiltrating immune cells can help to assess the anticancer response to immunotherapy in glioblastoma patients (50). 19 m6A regulators were highly expressed in glioma tissues (82). The expressions of m6A regulatory factors were related to the classification of glioma subtypes. The m6A modulators could predict prognosis and therapeutic effects, and were also related to the immune microenvironment of glioma (82, 83). The m6A modification regulator ELAVL1 was an effective predictor of PD-L1 treatment efficacy (83). Compared with normal brain tissues and glioblastoma tissues, most m6A RNA methylation regulators are differentially expressed in lower-grade gliomas tissues (85). Studies have revealed the correlation between TME infiltration of immune cells and m6A modification (86). In glioblastoma, WTAP was found to be overexpressed and to regulate tumor invasion and migration. High expression of WTAP was associated with a low postoperative survival rate. Furthermore, HNRNPC can also impact the invasiveness of glioblastoma cells and is regarded as a potential prognostic biomarker and therapeutic target for glioblastoma (86). Du et al. (87) comprehensively analyzed the m6A modification patterns of 1152 low-grade glioma samples, and found that the cases with a low m6A score had high immunogenicity and that those with a high m6A score were sensitive to chemo-radiotherapy and immunotherapy.
Digestive System Cancers
Gastric and Esophageal Cancers
Zhang et al. (88) reported that m6A modification plays an important role in the formation of TIME diversity and complexity by analyzing 21 m6A modulators in 1938 gastric cancer (GC) samples. Three m6A modification patterns including immune-excluded, immune-inflamed, and immune-desert phenotypes were discovered (88). m6A modification patterns could predict the stage of cancer-related inflammation, cancer subtypes, TME matrix activity, genetic variation, and patient prognosis (88–90). The high m6A-score subtype had a poor survival rate and had matrix activation with lack of effective immune infiltration; a low m6A score was associated with an increased neo-antigen load and an enhanced response to anti-PD-1/L1 immunotherapy (86–89). Assessing the m6A modification patterns of individual tumors will help to guide more effective immunotherapy strategies (88). High expression of WTAP was associated with RNA methylation, and its low expression was correlated with strong T cell-related immune responses in GC (86, 89). The infiltration of tumor-associated T cells in the TME was associated with high levels of m6A modification, which was mediated by WTAP mRNA expression (48, 89). Patients with high WTAP expression had fewer Tregs and CD4+ memory-activated T cells (48, 89). The high infiltration of Tregs and CD4+ memory-activated infiltrating T cells was associated with improved prognosis of GC patients (48, 89). Mo et al. (91) retrospectively analyzed 293 stomach adenocarcinoma samples using data from The Cancer Genome Atlas, and suggested that m6A methylation might also be used as an immunotherapy predictor in GCs.
The expressions of m6A modulators were correlated with the expressions of immuno-modulators and the level of immune infiltration in esophageal cancer, which can be divided into esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC) (92). The m6A modulators might improve the responsiveness of ESCC patients to immunotherapy by regulating the TIME and expression of PD-L1 (93).
Colorectal Cancer (CRC)
Based on the m6A signature score integrating m6A-related characteristic genes, patients with colon cancer (CC) could be divided into high- and low-score subgroups; a lower m6A score was associated with greater tumor mutation burden, higher PD-L1 expression, and higher SMG (such as PIK3CA and SMAD4) mutation rates (94, 95). The efficacy of immunotherapy for rectal cancer (RC) is closely related to the level of immune infiltration (96). Low expression of METTL14 in RC led to the downregulation of m6A RNA modification, which thereby reduced the level of immune cell infiltration and which led to a poor prognosis (96). The expression level of METTL14 was an independent prognostic factor in RC, and it was positively correlated with the level of immune infiltration. Durable neo-antigen-specific immunity was regulated by m6A RNA modification mediated by the m6A-binding protein YTHDF1 (56). In a CC-bearing mouse model, YTHDF1-deficient mice showed tumor growth inhibition and survived longer than wild-type mice (56). The loss of YTHDF1 in classical dendritic cells enhanced the cross-presentation of tumor antigens and the cross-priming of CD8+ T cells in vivo. In mice receiving anti-PD-L1 immunotherapy for CC, YTHDF1-deficient mice showed a higher cure rate (56). FTO was believed to regulate the methylation of PD-L1 mRNA thus determining the expression of PD-L1 in CC cells (97). Moreover, in CCs with proficient mismatch repair or low microsatellite instability, deletion of METTL3 and METTL14 increased the infiltration of CD8+ T cells and the levels of IFN-γ, Cxcl9, and Cxcl10 secretion and enhanced anti-PD-1 response (98, 99). CD34/CD276 affected the TIME and was modulated by m6A-dependent mechanisms, which ultimately promoted the immune escape of CRCs (100).
Hepatocellular Carcinoma (HCC)
Risk stratification by the expressions of 5 m6A-related genes (YTHDF1, HNRNPC, RBM15, METTL3, and YTHDF2) could improve the prognosis prediction of HCC and was related to the response to sorafenib treatment and anti-PD-1 immunotherapy (69, 101, 102). High expression of YTHDF2 was associated with poor prognosis of HCC, and together with increased immune cell infiltration, YTHDF2 might be an independent prognostic biomarker for HCC (103). HCC with low expression of METTL3 had increased dendritic cell infiltration in the TME (98). The levels of m6A methylation regulators were related to the overall survival and immunity in HCC, and METTL3, METTL13, YTHDF1, and YTHDF2 might be potential prognosis predictors and therapeutic targets in HCC (104, 105). Du et al. (106) used four m6A-related genes to construct a risk feature, which was associated with tumor immunity and which could stratify HCCs. m6A regulatory factors were significantly related to the TIME of HCC, which could divide HCCs into two clusters and which were associated with the expression level of programmed death ligand 1 (PD-L1), immune score, immune cell infiltration, and prognosis (107).
Pancreatic Cancer (PC)
Tang et al. (108) explored the correlation between M6A-related genes and the immune microenvironment of PC, and found that infiltrating immune cells might affect the M6A modification in tumor cells. An integrated model called “M6AScore” was constructed based on M6A modulation factors using RNA-seq data in pancreatic ductal adenocarcinoma-: M6AScore-high pancreatic ductal adenocarcinoma was characterized by immune reduction and T cell depletion, and M6AScore-low pancreatic ductal adenocarcinoma had higher reaction rates on immune checkpoint inhibitors (ICIs) treatment (109, 110). Thus, the M6AScore was associated with the invasiveness and immune status of PC, and could predict PC prognosis and response to ICIs treatment (108, 109, 111). Wang et al. (112) also established a prognostic model based on the expressions of m6A regulators, including IGF2BP2/3, KIAA1429, METTL3, EIF3H, and LRPPRC, which was associated with the TIME and immune statuses in PC.
Respiratory System Cancers
Lung Cancer (LC)
Lung adenocarcinoma is the most common histological manifestation of LC and is closely related to m6A abnormalities (95, 113–115). m6A methylation is reduced in the hyper-immune subtype of lung adenocarcinoma, indicating that m6A modification may mediate tumor immunity and provide potential anticancer therapeutic strategies (115). Compared with the low-risk lung squamous cell carcinoma patients, the expressions of ALKBH5, METL3, HNRNPC, and KIAA1429 were significantly reduced in patients with high-risk lung squamous cell carcinoma. It is worth noting that high-risk lung squamous cell carcinoma patients showed more promising treatment responses to PD-1 therapy (115). In non-small-cell lung cancer with high expressions of YTHDF1 and YTHDF2, the densities of four subsets of tumor-infiltrating lymphocytes (TILs; PD-1+, CD8+, Foxp3+, and CD45RO+) were significantly higher (116). High expressions of YTHDF1 and YTHDF2 were related to good prognosis of non-small-cell lung cancer patients, higher TIL density, and downregulation of PD-L1 (116).
Urinary System Cancers
Renal and Bladder Cancers
The expressions of FTO and METTL3 mRNAs were oppositely correlated with the expressions of CD8+ T cell migration-related chemokines in clear cell renal cell carcinoma (ccRCC), which might affect the antitumor immune response (117). Zhong et al. (118) constructed an m6A score to accurately evaluate the m6A methylation pattern in ccRCC patients, which could be used to predict the anti-PD-1 treatment response in ccRCC. In ccRCC the high m6A score group had higher PD-L1 expression, larger numbers of CD8+ T cells and CD4+ FOXP3+ Treg cells, and higher levels of immune cell infiltration (119). Among patients receiving immune checkpoint therapies, the clinical benefits were significantly higher in patients with high m6A scores (119). The expressions of 17 m6A RNA methylation regulators were closely related to the immunity and malignant progression of papillary renal cell carcinoma (120).
Overexpression of IL-32 is associated with m6A modification and good prognosis in bladder cancer, and which may promote the recruitment of CD4+ T cells and dendritic cells, thereby promoting the antitumor effect (121–124).
Reproductive System Cancers
Breast Cancer (BC)
He et al. (125) showed the significant association between RNA methylation levels and the numbers of tumor-infiltrating CD8+ T cells, regulatory T cells, helper T cells, activated NK cells, and M2 macrophages, which indicated the key roles of m6A modulators in the host anti-tumor immune response. Furthermore, the expression pattern of m6A modulators was also significantly related to the expression of PD-L1, TIM3, LAG3, and CCR4, which are well-known T cell depletion targets and important biomarkers in immunotherapy (126, 127) (126). The expression levels of METTL14 and ZC3H13 were significantly positively correlated with the infiltration levels of CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells, and negatively correlated with Treg cells in BC (128). BC genotypes could be divided into two clusters based on four representative m6A regulators (IGF2BP2, IGF2BP3, YTHDC2, and RBM15), which were associated with the number of TILs (129).
Ovarian, Endometrial, and Prostate Cancers
The levels of immune cell infiltration and various immune gene markers were closely related to the expressions of RBM15B, ZC3H13, YTHDF1, and IGF2BP1 in ovarian cancer (OC) (130). Gu et al. (131) identified two different m6A modes based on 21 m6A regulators: A low m6A score was associated with immune activation and stronger sensitivity to immune checkpoint inhibitors, while a high m6A score was associated with progression of OC.
The expressions of METTL14, ZC3H13, and YTHDC1 were positively correlated with the expression of PD-L1 in endometrial cancer (EC) (132). Knockdown of ZC3H13 or YTHDC1 in vitro promoted the malignant phenotype transformation of EC cells.
The expressions of HNRNPA2B1 and METTL3 may also affect the immune microenvironment of prostate cancer (133).
Hematologic System Cancers
Acute Myeloid Leukemia (AML)
Acute myeloid leukemia (AML) is a blood cancer that affects a specific subgroup of hematopoietic stem/progenitor cells, and has different genetic and molecular abnormalities (105, 126, 127, 134). In AML, YTHDF2 could isolate m6A-modified circRNA and inhibit innate immunity (125). For unmodified circRNA, it could be used as an effective adjuvant to induce antigen-specific T cell activation, antibody production, and antitumor immunity enhancement (125). The genetic depletion and pharmacological inhibition of FTO significantly inhibited the self-renewal of leukemia stem cells, and induced immune responses by inhibiting the expression of immune checkpoint genes (128). Targeting FTO could make leukemia cells more sensitive to T cell toxicity and overcome immune evasion induced by hypo-methylation agents, suggesting the potential value of targeting FTO in anticancer treatment (128).
Other Cancers
Knockdown of FTO increased the m6A methylation of key tumor-promoting genes in melanoma cells, including PD-1, CXCR4, and SOX10, resulting in increased RNA attenuation mediated by the m6A reader YTHDF2 (129, 135, 136). Knockdown of FTO made melanoma cells more sensitive to interferon gamma (IFN-γ), thereby reducing the resistance to anti-PD-1 therapy in mice in an adaptive immunity-dependent manner (129, 137). The combination of FTO inhibition and anti-PD-1 blockade reduced the resistance of melanoma to immunotherapy (86). The expression and mutation statuses of the ALKBH5 gene were closely related to the response to immunotherapy in patients with melanoma (75). Knockout of ALKBH5 in tumor cells enhanced the efficacy of immunotherapy, which supported the therapeutic value of ALKBH5 in melanoma immunotherapy (75). Mutation or downregulation of the ALKBH5 gene in melanoma patients was associated with positive response to PD-1 blockade by pembrolizumab or nivolumab (75).
m6A RNA methylation may be involved in the regulation of the immune microenvironment in head and neck squamous cell carcinoma (HNSCC) in synergy with the PI3K/AKT/mTOR signaling pathway (138). Li et al. found that YTHDC2 is associated with the level of immune infiltration of B cells, CD8+ T cells, CD4+ T cells, neutrophils, and dendritic cells in HNSCC (139). Feng et al. (140) further revealed the important role of m6A RNA methylation-related lncRNAs in the HNSCC immune microenvironment. Nasopharyngeal carcinoma (NPC) is a highly immunogenic tumor, which is characterized by a large abundance of tumor infiltrating lymphocytes. METTL3 was low expressed in NPC and related to the infiltration of various immune cells (76, 141–144).
Adrenocortical carcinoma (ACC) is a highly immunogenic tumor, and 86.3% of ACCs had abundant tumor infiltrating lymphocytes (145). The m6A reader HNRNPA2B1 mediated the pattern of TME infiltration, and promoted the progression of ACC by regulating the activity of macrophages (145).
Numbers of most of the immune cells (type M1 and M2 macrophages, CD8+ T cells, Tregs, and dendritic cells) were negatively associated with IGF2BP2 expression in osteosarcoma (146). M6A modification-mediated aberrant activation of cell cycle-related pathways and suppression of immune response may play a crucial role in the progression of osteosarcoma (147).
Perspectives
m6A RNA methylation modification plays various vital roles in nearly all biology processes including cancer initiation and progression. There are some unsolved questions that need to be addressed in the future to fully reveal the function of m6A modification during tumor genesis, progression, and antitumor immune response. The roles of m6A methylation need to be studied in more types of immune cells and immune-associated cells besides those herein reviewed, and the involvement of m6A modification in the regulation of more biology behaviors and functions (e.g., metabolism) of immune cells and in the interplay and crosstalk between cancer cells, immune cells, other stromal cells, and non-cellular TME components need to be further investigated, to comprehensively reveal the complicated m6A-associated regulatory networks and to provide promising targets for novel immunotherapy strategies. The m6A-modified molecules and the relevant signaling pathways which have been investigated are limited, and the full m6A regulatory spectra within specific immune cells need to be further uncovered. Notably, the effects of m6A methylation on the biology behavior and function of a certain type of immune cell may vary according to different target RNAs (mRNAs and non-coding RNAs), and the interaction between different regulatory pathways need to be clarified. Besides the extracellular m6A inducers (e.g., IL-6 and lipopolysaccharide) herein reviewed, explorations of other inducers and inhibitors may offer better understanding of the meticulous m6A regulatory network. The m6A regulators which have currently been explored are also limited; deciphering the roles of more m6A writers, erasers, and readers and the further interplays between them and identification of the most relevant and potent regulators may provide important hints for development of m6A-relevant targets for precision immunotherapy. While the inhibition of YTHDF1 and/or YTHDF2 in DCs and macrophages have been suggested to be promising anticancer strategy, the influences of such inhibition on the other intracellular molecules and pathways besides those already explored and on cancer cells, other immune cells, and other stromal cells also need to be clarified to further support the therapeutic significance with safety; it would also be important to explore how to precisely deliver the corresponding inhibitory drugs to the specific cell types to minimize the off-target effects. It may also be interesting to investigate the impact of m6A modification on RNA in different organelles, such as mitochondria and extracellular vesicles. The different regulatory roles of m6A methylation in different stages of tumor progression and in primary and metastatic cancer sites and the interplay between m6A and other RNA modifications may also be intriguing topics of research.
Most of the currently available evidence on the roles of m6A modification in the regulation of tumor immunity in the TME can be divided into two aspects: Exploration of the biology function of a single m6A modulator to clarify the underlying mechanism and construction of model or signature integrating multiple m6A regulators to precisely predict the prognosis and immunotherapy efficacy within a specific cancer. For mechanism explorations, it is encouraged that the impact of m6A modification on more tumor biology behaviors including immune metabolism, extracellular vesicular activity, and autophagy beyond the routinely investigated ones be evaluated. It is important to also look more into the feedback modulation of m6A modification by the TIME, and a feedback loop between m6A methylation and tumor immunity may better represent the real biology processes within humans. A target RNA is regulated by multiple m6A modulators, and a modulator can function on various target RNAs. The interaction between diverse regulatory pathways cannot be neglected, and artificial intelligence (AI) and bioinformatics methods may help to better understand the complex processes. While various m6A-based signatures or scores have been constructed, they are mostly based on publicly available databases (e.g., TCGA) which include mostly western populations and often lack validations in different ethnicities. It is warranted that more precision models will be derived from clinically oriented data within different populations and be validated across countries or continents, to largely improve the representativeness and generalizability. The methods used to build the m6A-based models are also largely heterogeneous across studies. Comparative studies paralleling investigating the prediction efficacy of models constructed using different methods, including deep learning, other machine learning (e.g., the classical support vector machine), and the popular LASSO regression for selecting more predictively meaningful variables, may help to identify the best model.
Previously, using machine learning, we have constructed two personalized signatures majorly based on multiple selected immune cell markers in the TME which can precisely predict prognosis and chemotherapy benefits for a specific patient (148–150); furthermore, we also build an individualized predictive model based on immune cells in the peripheral blood to help to precisely select the subgroup of patients who will more likely benefit from radical resection (151). It may be interesting to investigate the associations between m6A methylation and tumor immunity-based signatures or scores. For m6A-based model construction, it is investigable to alternatively change the endpoint from survival outcomes to signatures summarizing characteristics of the TIME, and the models constructed may also be well predictive of longer-term outcomes. Since tissue specimens are not always obtainable especially among patients with advanced or metastatic cancers that are not resectable, it is necessary to fully consider the concept of liquid biopsy and to find surrogate markers within easily accessible samples, especially the peripheral blood. It is encouraged that further studies focus on the association between the m6A regulators and tumor immunity markers in the cancer lesion and in easily accessible body liquids to clarify the representativeness of the latter, and then on the clinical usefulness of m6A-based models constructed based on the peripheral blood markers. It may also be interesting to assess the efficacy of predicting the previously constructive immune signature and then prognosis and treatment benefits based on scores integrating m6A regulators, and models integrating both the m6A regulators and tumor immunity components may have further enhanced predictive efficacy.
Based on the close relationship between m6A modification and tumor immunity, m6A-targeted immunotherapy strategies may be promising. Regulations of the methylation and demethylation statuses within diverse immune cells may dramatically impact the antitumor activities, and rebalancing the homeostasis of m6A writers, erasers, and readers may contribute to creating a favorable anticancer immunity effectively inhibiting tumor initiation and progression. m6A-based signatures can well predict the efficacy of immunotherapy using agents targeting PD-1/L1, etc., and it is desirable that m6A-targeted therapeutic strategies will further enhance the efficacy of immunotherapy; targeting both tumor immunity and m6A modification may have markedly stronger effects compared to targeting either alone. It may be recommendable that m6A modulators associated with anticancer immune components and activities be selected and form the basis for further drug development; particularly, those with clear and obvious interactions with famous cancer immune markers whose functions have been well revealed (e.g., PD-1/L1, CTLA4, and CD47) should be the focus. Since m6A modification is a relatively new concept, there may still be some way to go for the development of relevant agents. Notably, there are complex m6A regulatory networks accompanied by numerous feedback loops and other precise regulatory mechanisms within human bodies, and targeting a single modulator may not fully produce the anticipated effects. Action on multiple m6A modulators to reshape the relevant homeostasis may have better anticancer effects. Furthermore, to minimize the possible off-target effects, it may be desirable to more precisely deliver the m6A-targeted modulatory agents into the immune cells of interest, with the help of some advanced drug delivery technologies (e.g., nanotechnology). Importantly, it is unclear whether change in level of a specific modulator will cause a dramatic cascade reaction greatly impairing other body functioning, and the influences of the “butterfly flapping its wings” should be carefully assessed before any m6A-targeted agents are used in humans.
Conclusions
This review focuses on the modulation of immune function by m6A in the TME of various cancers. m6A is one of the most common RNA modifications and affects tumor occurrence, development, and response to immunotherapy by regulating anticancer immunity. The m6A writers, erasers, and readers, which are involved in nearly all biology processes of RNA, such as maturation, transport, splicing, translation, and degradation, all play important roles in the modulation of anticancer immune response. m6A can impact anticancer immunity by regulating diverse activities of various immune cells, such as the differentiation of T cells, the stabilization of Tregs, the maturation of DCs, the polarization of macrophages, and the function modulation of MDSCs. m6A modulators are closely related to tumor immunity and immunotherapy, and many abnormally expressed m6A regulatory factors can impact anticancer immune functions and further modulate tumor genesis, proliferation, growth, invasion, and metastasis by regulating the balance between the expressions of oncogenes and tumor-suppressor genes in the TIME. Notably, most of the reviewed studies on the relationship between m6A RNA methylation and tumor immunity are still in their infancy, and more in-depth researches are needed to explore the mechanisms underlying the regulation of tumor immunity by m6A modifications.
Author Contributions
Conception or design: LG, YS, and LH. Literature retrieval and review: LG and LH. Drafting of the manuscript: LG and LH. Critical revision of the manuscript for important intellectual content: HY, CZ, YS, LH, and JZ. Administrative, technical, or material support: YS and JZ. All authors contributed to the article and approved the submitted version.
Funding
This study was sponsored by Shanghai Pujiang Program (21PJ1409700) and by the Start-up Fund for the Introduction of High Level Talents by Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. The funders had no involvement in study design; in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dai D, Wang H, Zhu L, Jin H, Wang X. N6-Methyladenosine Links RNA Metabolism to Cancer Progression. Cell Death Dis (2018) 9(2):124. doi: 10.1038/s41419-017-0129-x
2. Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM, et al. N (6)-Methyladenosine of Chromosome-Associated Regulatory RNA Regulates Chromatin State and Transcription. Science (2020) 367(6477):580–6. doi: 10.1126/science.aay6018
3. Jia C, Wang G, Wang T, Fu B, Zhang Y, Huang L, et al. Cancer-Associated Fibroblasts Induce Epithelial-Mesenchymal Transition via the Transglutaminase 2-Dependent IL-6/IL6R/STAT3 Axis in Hepatocellular Carcinoma. Int J Biol Sci (2020) 16(14):2542–58. doi: 10.7150/ijbs.45446
4. Chen XY, Zhang J, Zhu JS. The Role of M(6)A RNA Methylation in Human Cancer. Mol Cancer (2019) 18(1):103. doi: 10.1186/s12943-019-1033-z
5. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The Interplay Between M6a RNA Methylation and Noncoding RNA in Cancer. J Hematol Oncol (2019) 12(1):121. doi: 10.1186/s13045-019-0805-7
6. Liu ZX, Li LM, Sun HL, Liu SM. Link Between M6a Modification and Cancers. Front Bioeng Biotechnol (2018) 6:89. doi: 10.3389/fbioe.2018.00089
7. Surendran V, Rutledge D, Colmon R, Chandrasekaran A. A Novel Tumor-Immune Microenvironment (TIME)-On-Chip Mimics Three Dimensional Neutrophil-Tumor Dynamics and Neutrophil Extracellalar Traps (NETs)- Mediated Collective Tumor Invasion. Biofabrication (2021). doi: 10.1088/1758-5090/abe1cf
8. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the Tumor Immune Microenvironment (TIME) for Effective Therapy. Nat Med (2018) 24(5):541–50. doi: 10.1038/s41591-018-0014-x
9. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural Basis of N(6)-Adenosine Methylation by the METTL3-METTL14 Complex. Nature (2016) 534(7608):575–8. doi: 10.1038/nature18298
10. Ma Z, Gao X, Shuai Y, Xing X, Ji J. The M6a Epitranscriptome Opens a New Charter in Immune System Logic. Epigenetics (2021) 16(8):819–37. doi: 10.1080/15592294.2020.1827722
11. Shulman Z, Stern-Ginossar N. The RNA Modification N(6)-Methyladenosine as a Novel Regulator of the Immune System. Nat Immunol (2020) 21(5):501–12. doi: 10.1038/s41590-020-0650-4
12. Zhao Y, Shi Y, Shen H, Xie W. M(6)A-Binding Proteins: The Emerging Crucial Performers in Epigenetics. J Hematol Oncol (2020) 13(1):35. doi: 10.1186/s13045-020-00872-8
13. Yang C, Hu Y, Zhou B, Bao Y, Li Z, Gong C, et al. The Role of M(6)A Modification in Physiology and Disease. Cell Death Dis (2020) 11(11):960. doi: 10.1038/s41419-020-03143-z
14. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-Methyladenosine and Its Role in Cancer. Mol Cancer (2019) 18(1):176. doi: 10.1186/s12943-019-1109-9
15. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. M(6)A RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature (2016) 537(7620):369–73. doi: 10.1038/nature19342
16. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of M6a Writers Reveals Two Distinct Classes of mRNA Methylation at Internal and 5’ Sites. Cell Rep (2014) 8(1):284–96. doi: 10.1016/j.celrep.2014.05.048
17. Mendel M, Chen KM, Homolka D, Gos P, Pandey RR, McCarthy AA, et al. Methylation of Structured RNA by the M(6)A Writer METTL16 Is Essential for Mouse Embryonic Development. Mol Cell (2018) 71(6):986–1000 e11. doi: 10.1016/j.molcel.2018.08.004
18. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA M(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell (2017) 169(5):824–35 e14. doi: 10.1016/j.cell.2017.05.003
19. Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Hobartner C, et al. Human METTL16 Is a N(6)-Methyladenosine (M(6)A) Methyltransferase That Targets pre-mRNAs and Various Non-Coding RNAs. EMBO Rep (2017) 18(11):2004–14. doi: 10.15252/embr.201744940
20. Yuan S, Tang H, Xing J, Fan X, Cai X, Li Q, et al. Methylation by NSun2 Represses the Levels and Function of microRNA 125b. Mol Cell Biol (2014) 34(19):3630–41. doi: 10.1128/MCB.00243-14
21. Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R, et al. N(6-)Methyladenosine Methyltransferase ZCCHC4 Mediates Ribosomal RNA Methylation. Nat Chem Biol (2019) 15(1):88–94. doi: 10.1038/s41589-018-0184-3
22. Liang Z, Kidwell RL, Deng H, Xie Q. Epigenetic N6-Methyladenosine Modification of RNA and DNA Regulates Cancer. Cancer Biol Med (2020) 17(1):9–19. doi: 10.20892/j.issn.2095-3941.2019.0347
23. Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D, et al. Mechanism of RNA Modification N6-Methyladenosine in Human Cancer. Mol Cancer (2020) 19(1):104. doi: 10.1186/s12943-020-01216-3
24. Wu B, Li L, Huang Y, Ma J, Min J. Readers, Writers and Erasers of N(6)-Methylated Adenosine Modification. Curr Opin Struct Biol (2017) 47:67–76. doi: 10.1016/j.sbi.2017.05.011
25. Meyer KD, Jaffrey SR. Rethinking M(6)A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol (2017) 33:319–42. doi: 10.1146/annurev-cellbio-100616-060758
26. Sivasudhan E, Blake N, Lu ZL, Meng J, Rong R. Dynamics of M6a RNA Methylome on the Hallmarks of Hepatocellular Carcinoma. Front Cell Dev Biol (2021) 9:642443. doi: 10.3389/fcell.2021.642443
27. Zhou KI, Pan T. An Additional Class of M(6)A Readers. Nat Cell Biol (2018) 20(3):230–2. doi: 10.1038/s41556-018-0046-y
28. Nombela P, Miguel-Lopez B, Blanco S. The Role of M(6)A, M(5)C and Psi RNA Modifications in Cancer: Novel Therapeutic Opportunities. Mol Cancer (2021) 20(1):18. doi: 10.1186/s12943-020-01263-w
29. He L, Li J, Wang X, Ying Y, Xie H, Yan H, et al. The Dual Role of N6-Methyladenosine Modification of RNAs Is Involved in Human Cancers. J Cell Mol Med (2018) 22(10):4630–9. doi: 10.1111/jcmm.13804
30. Berger Y, Giurcanu M, Vining CC, Schuitevoerder D, Posner MC, Roggin KK, et al. Cytoreductive Surgery for Selected Patients Whose Metastatic Gastric Cancer was Treated With Systemic Chemotherapy. Ann Surg Oncol (2021) 28(8):4433–3. doi: 10.1245/s10434-020-09475-6
31. Shi H, Wei J, He C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol Cell (2019) 74(4):640–50. doi: 10.1016/j.molcel.2019.04.025
32. Wang YN, Yu CY, Jin HZ. RNA N(6)-Methyladenosine Modifications and the Immune Response. J Immunol Res (2020) 2020:6327614. doi: 10.1155/2020/6327614
33. Huang H, Weng H, Chen J. M(6)A Modification in Coding and Non-Coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell (2020) 37(3):270–88. doi: 10.1016/j.ccell.2020.02.004
34. Zhang Y, Geng X, Li Q, Xu J, Tan Y, Xiao M, et al. M6a Modification in RNA: Biogenesis, Functions and Roles in Gliomas. J Exp Clin Cancer Res (2020) 39(1):192. doi: 10.1186/s13046-020-01706-8
35. Wang Y, Zhang Y, Du Y, Zhou M, Hu Y, Zhang S. Emerging Roles of N6-Methyladenosine (M(6)A) Modification in Breast Cancer. Cell Biosci (2020) 10(1):136. doi: 10.1186/s13578-020-00502-3
36. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the Human and Mouse M6a RNA Methylomes Revealed by M6a-Seq. Nature (2012) 485(7397):201–6. doi: 10.1038/nature11112
37. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3’ UTRs and Near Stop Codons. Cell (2012) 149(7):1635–46. doi: 10.1016/j.cell.2012.05.003
38. Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-Nucleotide-Resolution Mapping of M6a and m6Am Throughout the Transcriptome. Nat Methods (2015) 12(8):767–72. doi: 10.1038/nmeth.3453
39. Chen Y, Lin Y, Shu Y, He J, Gao W. Interaction Between N(6)-Methyladenosine (M(6)A) Modification and Noncoding RNAs in Cancer. Mol Cancer (2020) 19(1):94. doi: 10.1186/s12943-020-01207-4
40. Gu Y, Wu X, Zhang J, Fang Y, Pan Y, Shu Y, et al. The Evolving Landscape of N(6)-Methyladenosine Modification in the Tumor Microenvironment. Mol Ther (2021) 29(5):1703–15. doi: 10.1016/j.ymthe.2021.04.009
41. Wu K, Lin K, Li X, Yuan X, Xu P, Ni P, et al. Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment. Front Immunol (2020) 11:1731. doi: 10.3389/fimmu.2020.01731
42. Si C, Chen C, Guo Y, Kang Q, Sun Z. Effect, Mechanism, and Applications of Coding/Non-Coding RNA M6a Modification in Tumor Microenvironment. Front Cell Dev Biol (2021) 9:711815. doi: 10.3389/fcell.2021.711815
43. Barker HE, Paget JT, Khan AA, Harrington KJ. The Tumour Microenvironment After Radiotherapy: Mechanisms of Resistance and Recurrence. Nat Rev Cancer (2015) 15(7):409–25. doi: 10.1038/nrc3958
44. Li M, Zha X, Wang S. The Role of N6-Methyladenosine mRNA in the Tumor Microenvironment. Biochim Biophys Acta Rev Cancer (2021) 1875:188522. doi: 10.1016/j.bbcan.2021.188522
45. Zhu J, Xiao J, Wang M, Hu D. Pan-Cancer Molecular Characterization of M(6)A Regulators and Immunogenomic Perspective on the Tumor Microenvironment. Front Oncol (2020) 10:618374. doi: 10.3389/fonc.2020.618374
46. Lou X, Wang JJ, Wei YQ, Sun JJ. Emerging Role of RNA Modification N6-Methyladenosine in Immune Evasion. Cell Death Dis (2021) 12(4):300. doi: 10.1038/s41419-021-03585-z
47. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. M(6)A mRNA Methylation Controls T Cell Homeostasis by Targeting the IL-7/STAT5/SOCS Pathways. Nature (2017) 548(7667):338–42. doi: 10.1038/nature23450
48. Zhang M, Song J, Yuan W, Zhang W, Sun Z. Roles of RNA Methylation on Tumor Immunity and Clinical Implications. Front Immunol (2021) 12:641507. doi: 10.3389/fimmu.2021.641507
49. Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6-Methyladenosine Methyltransferase-Like 3 Promotes Liver Cancer Progression Through YTHDF2-Dependent Posttranscriptional Silencing of SOCS2. Hepatology (2018) 67(6):2254–70. doi: 10.1002/hep.29683
50. Tong J, Cao G, Zhang T, Sefik E, Amezcua Vesely MC, Broughton JP, et al. M(6)A mRNA Methylation Sustains Treg Suppressive Functions. Cell Res (2018) 28(2):253–6. doi: 10.1038/cr.2018.7
51. Li M, Zha X, Wang S. The Role of N6-Methyladenosine mRNA in the Tumor Microenvironment. Biochim Biophys Acta Rev Cancer (2021) 1875(2):188522. doi: 10.1016/j.bbcan.2021.188522
52. Gu J, Zhan Y, Zhuo L, Zhang Q, Li G, Li Q, et al. Biological Functions of M(6)A Methyltransferases. Cell Biosci (2021) 11(1):15. doi: 10.1186/s13578-020-00513-0
53. Eisenbarth SC. Dendritic Cell Subsets in T Cell Programming: Location Dictates Function. Nat Rev Immunol (2019) 19(2):89–103. doi: 10.1038/s41577-018-0088-1
54. Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, et al. Mettl3-Mediated mRNA M(6)A Methylation Promotes Dendritic Cell Activation. Nat Commun (2019) 10(1):1898. doi: 10.1038/s41467-019-09903-6
55. Feng Z, Li Q, Meng R, Yi B, Xu Q. METTL3 Regulates Alternative Splicing of MyD88 Upon the Lipopolysaccharide-Induced Inflammatory Response in Human Dental Pulp Cells. J Cell Mol Med (2018) 22(5):2558–68. doi: 10.1111/jcmm.13491
56. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-Tumour Immunity Controlled Through mRNA M(6)A Methylation and YTHDF1 in Dendritic Cells. Nature (2019) 566(7743):270–4. doi: 10.1038/s41586-019-0916-x
57. Alloatti A, Kotsias F, Magalhaes JG, Amigorena S. Dendritic Cell Maturation and Cross-Presentation: Timing Matters! Immunol Rev (2016) 272(1):97–108. doi: 10.1111/imr.12432
58. Cebrian I, Visentin G, Blanchard N, Jouve M, Bobard A, Moita C, et al. Sec22b Regulates Phagosomal Maturation and Antigen Crosspresentation by Dendritic Cells. Cell (2011) 147(6):1355–68. doi: 10.1016/j.cell.2011.11.021
59. Burbage M, Gros M, Amigorena S. Translate Less, Prime Better, to Improve Anti-Tumor Responses. Nat Immunol (2019) 20(5):518–20. doi: 10.1038/s41590-019-0371-8
60. Roberts EW, Broz ML, Binnewies M, Headley MB, Nelson AE, Wolf DM, et al. Critical Role for CD103(+)/CD141(+) Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell (2016) 30(2):324–36. doi: 10.1016/j.ccell.2016.06.003
61. Liu J, Zhang X, Chen K, Cheng Y, Liu S, Xia M, et al. CCR7 Chemokine Receptor-Inducible lnc-Dpf3 Restrains Dendritic Cell Migration by Inhibiting HIF-1alpha-Mediated Glycolysis. Immunity (2019) 50(3):600–15 e15. doi: 10.1016/j.immuni.2019.01.021
62. Zhou J, Tang Z, Gao S, Li C, Feng Y, Zhou X. Tumor-Associated Macrophages: Recent Insights and Therapies. Front Oncol (2020) 10:188. doi: 10.3389/fonc.2020.00188
63. Lavin Y, Mortha A, Rahman A, Merad M. Regulation of Macrophage Development and Function in Peripheral Tissues. Nat Rev Immunol (2015) 15(12):731–44. doi: 10.1038/nri3920
64. Gu X, Zhang Y, Li D, Cai H, Cai L, Xu Q. N6-Methyladenosine Demethylase FTO Promotes M1 and M2 Macrophage Activation. Cell Signal (2020) 69:109553. doi: 10.1016/j.cellsig.2020.109553
65. Chen J, Du B. Novel Positioning From Obesity to Cancer: FTO, an M(6)A RNA Demethylase, Regulates Tumour Progression. J Cancer Res Clin Oncol (2019) 145(1):19–29. doi: 10.1007/s00432-018-2796-0
66. Li C, Xu MM, Wang K, Adler AJ, Vella AT, Zhou B. Macrophage Polarization and Meta-Inflammation. Transl Res (2018) 191:29–44. doi: 10.1016/j.trsl.2017.10.004
67. Shrivastava R, Asif M, Singh V, Dubey P, Ahmad Malik S, Lone MU, et al. M2 Polarization of Macrophages by Oncostatin M in Hypoxic Tumor Microenvironment Is Mediated by Mtorc2 and Promotes Tumor Growth and Metastasis. Cytokine (2019) 118:130–43. doi: 10.1016/j.cyto.2018.03.032
68. Liu Y, Liu Z, Tang H, Shen Y, Gong Z, Xie N, et al. The N(6)-Methyladenosine (M(6)A)-Forming Enzyme METTL3 Facilitates M1 Macrophage Polarization Through the Methylation of STAT1 mRNA. Am J Physiol Cell Physiol (2019) 317(4):C762–C75. doi: 10.1152/ajpcell.00212.2019
69. He Y, Xing J, Wang S, Xin S, Han Y, Zhang J. Increased M6a Methylation Level Is Associated With the Progression of Human Abdominal Aortic Aneurysm. Ann Transl Med (2019) 7(24):797. doi: 10.21037/atm.2019.12.65
70. Huangfu N, Zheng W, Xu Z, Wang S, Wang Y, Cheng J, et al. RBM4 Regulates M1 Macrophages Polarization Through Targeting STAT1-Mediated Glycolysis. Int Immunopharmacol (2020) 83:106432. doi: 10.1016/j.intimp.2020.106432
71. Veglia F, Perego M, Gabrilovich D. Myeloid-Derived Suppressor Cells Coming of Age. Nat Immunol (2018) 19(2):108–19. doi: 10.1038/s41590-017-0022-x
72. Ni HH, Zhang L, Huang H, Dai SQ, Li J. Connecting METTL3 and Intratumoural CD33(+) MDSCs in Predicting Clinical Outcome in Cervical Cancer. J Transl Med (2020) 18(1):393. doi: 10.1186/s12967-020-02553-z
73. Law AMK, Valdes-Mora F, Gallego-Ortega D. Myeloid-Derived Suppressor Cells as a Therapeutic Target for Cancer. Cells (2020) 9(3):561. doi: 10.3390/cells9030561
74. Shang W, Gao Y, Tang Z, Zhang Y, Yang R. The Pseudogene Olfr29-Ps1 Promotes the Suppressive Function and Differentiation of Monocytic MDSCs. Cancer Immunol Res (2019) 7(5):813–27. doi: 10.1158/2326-6066.CIR-18-0443
75. Li N, Kang Y, Wang L, Huff S, Tang R, Hui H, et al. ALKBH5 Regulates Anti-PD-1 Therapy Response by Modulating Lactate and Suppressive Immune Cell Accumulation in Tumor Microenvironment. Proc Natl Acad Sci USA (2020) 117(33):20159–70. doi: 10.1073/pnas.1918986117
76. Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, et al. Promoter-Bound METTL3 Maintains Myeloid Leukaemia by M(6)A-Dependent Translation Control. Nature (2017) 552(7683):126–31. doi: 10.1038/nature24678
77. Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. M(6)A mRNA Methylation Regulates AKT Activity to Promote the Proliferation and Tumorigenicity of Endometrial Cancer. Nat Cell Biol (2018) 20(9):1074–83. doi: 10.1038/s41556-018-0174-4
78. Galon J, Bruni D. Tumor Immunology and Tumor Evolution: Intertwined Histories. Immunity (2020) 52(1):55–81. doi: 10.1016/j.immuni.2019.12.018
79. Tang L, Wei X, Li T, Chen Y, Dai Z, Lu C, et al. Emerging Perspectives of RNA N (6)-Methyladenosine (M(6)A) Modification on Immunity and Autoimmune Diseases. Front Immunol (2021) 12:630358. doi: 10.3389/fimmu.2021.630358
80. Newton JM, Hanoteau A, Liu HC, Gaspero A, Parikh F, Gartrell-Corrado RD, et al. Immune Microenvironment Modulation Unmasks Therapeutic Benefit of Radiotherapy and Checkpoint Inhibition. J Immunother Cancer (2019) 7(1):216. doi: 10.1186/s40425-019-0698-6
81. Qu S, Chen Z, Liu B, Liu J, Wang H. N6-Methyladenine-Related Genes Affect Biological Behavior and the Prognosis of Glioma. Cancer Med (2021) 10(1):98–108. doi: 10.1002/cam4.3574
82. Xu S, Tang L, Dai G, Luo C, Liu Z. Expression of M6a Regulators Correlated With Immune Microenvironment Predicts Therapeutic Efficacy and Prognosis in Gliomas. Front Cell Dev Biol (2020) 8:594112. doi: 10.3389/fcell.2020.594112
83. Pan Y, Xiao K, Li Y, Li Y, Liu Q. RNA N6-Methyladenosine Regulator-Mediated Methylation Modifications Pattern and Immune Infiltration Features in Glioblastoma. Front Oncol (2021) 11:632934. doi: 10.3389/fonc.2021.632934
84. Pinello N, Sun S, Wong JJ. Aberrant Expression of Enzymes Regulating M(6)A mRNA Methylation: Implication in Cancer. Cancer Biol Med (2018) 15(4):323–34. doi: 10.20892/j.issn.2095-3941.2018.0365
85. Zheng J, Wang X, Qiu Y, Wang M, Yu H, Zhou Z, et al. Identification of Critical M(6)A RNA Methylation Regulators With Prognostic Value in Lower-Grade Glioma. BioMed Res Int (2021) 2021:9959212. doi: 10.1155/2021/9959212
86. Lin S, Xu H, Zhang A, Ni Y, Xu Y, Meng T, et al. Prognosis Analysis and Validation of M(6)A Signature and Tumor Immune Microenvironment in Glioma. Front Oncol (2020) 10:541401. doi: 10.3389/fonc.2020.541401
87. Du J, Ji H, Ma S, Jin J, Mi S, Hou K, et al. M6a Regulator-Mediated Methylation Modification Patterns and Characteristics of Immunity and Stemness in Low-Grade Glioma. Brief Bioinform (2021) 22(5):bbab013. doi: 10.1093/bib/bbab013 PubMed PMID: 33594424.
88. Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. M(6)A Regulator-Mediated Methylation Modification Patterns and Tumor Microenvironment Infiltration Characterization in Gastric Cancer. Mol Cancer (2020) 19(1):53. doi: 10.1186/s12943-020-01170-0
89. Li H, Su Q, Li B, Lan L, Wang C, Li W, et al. High Expression of WTAP Leads to Poor Prognosis of Gastric Cancer by Influencing Tumour-Associated T Lymphocyte Infiltration. J Cell Mol Med (2020) 24(8):4452–65. doi: 10.1111/jcmm.15104
90. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-Mediated M(6)A Modification of HDGF mRNA Promotes Gastric Cancer Progression and has Prognostic Significance. Gut (2020) 69(7):1193–205. doi: 10.1136/gutjnl-2019-319639
91. Mo P, Xie S, Cai W, Ruan J, Du Q, Ye J, et al. N(6)-Methyladenosine (M(6)A) RNA Methylation Signature as a Predictor of Stomach Adenocarcinoma Outcomes and its Association With Immune Checkpoint Molecules. J Int Med Res (2020) 48(9):300060520951405. doi: 10.1177/0300060520951405
92. Zhao H, Xu Y, Xie Y, Zhang L, Gao M, Li S, et al. M6a Regulators Is Differently Expressed and Correlated With Immune Response of Esophageal Cancer. Front Cell Dev Biol (2021) 9:650023. doi: 10.3389/fcell.2021.650023
93. Guo W, Tan F, Huai Q, Wang Z, Shao F, Zhang G, et al. Comprehensive Analysis of PD-L1 Expression, Immune Infiltrates, and M6a RNA Methylation Regulators in Esophageal Squamous Cell Carcinoma. Front Immunol (2021) 12:669750. doi: 10.3389/fimmu.2021.669750
94. Chong W, Shang L, Liu J, Fang Z, Du F, Wu H, et al. M(6)A Regulator-Based Methylation Modification Patterns Characterized by Distinct Tumor Microenvironment Immune Profiles in Colon Cancer. Theranostics (2021) 11(5):2201–17. doi: 10.7150/thno.52717
95. Liu T, Yang S, Sui J, Xu SY, Cheng YP, Shen B, et al. Dysregulated N6-Methyladenosine Methylation Writer METTL3 Contributes to the Proliferation and Migration of Gastric Cancer. J Cell Physiol (2020) 235(1):548–62. doi: 10.1002/jcp.28994
96. Cai C, Long J, Huang Q, Han Y, Peng Y, Guo C, et al. M6A “Writer” Gene METTL14: A Favorable Prognostic Biomarker and Correlated With Immune Infiltrates in Rectal Cancer. Front Oncol (2021) 11:615296. doi: 10.3389/fonc.2021.615296
97. Tsuruta N, Tsuchihashi K, Ohmura H, Yamaguchi K, Ito M, Ariyama H, et al. RNA N6-Methyladenosine Demethylase FTO Regulates PD-L1 Expression in Colon Cancer Cells. Biochem Biophys Res Commun (2020) 530(1):235–9. doi: 10.1016/j.bbrc.2020.06.153
98. Wang L, Hui H, Agrawal K, Kang Y, Li N, Tang R, et al. M(6) A RNA Methyltransferases METTL3/14 Regulate Immune Responses to Anti-PD-1 Therapy. EMBO J (2020) 39(20):e104514. doi: 10.15252/embj.2020104514
99. Xu X, Huang J, Ocansey DKW, Xia Y, Zhao Z, Xu Z, et al. The Emerging Clinical Application of M6a RNA Modification in Inflammatory Bowel Disease and Its Associated Colorectal Cancer. J Inflammation Res (2021) 14:3289–306. doi: 10.2147/JIR.S320449
100. Zhou Y, Zhou H, Shi J, Guan A, Zhu Y, Hou Z, et al. Decreased M6a Modification of CD34/CD276(B7-H3) Leads to Immune Escape in Colon Cancer. Front Cell Dev Biol (2021) 9:715674. doi: 10.3389/fcell.2021.715674
101. Su Y, Huang J, Hu J. M(6)A RNA Methylation Regulators Contribute to Malignant Progression and Have Clinical Prognostic Impact in Gastric Cancer. Front Oncol (2019) 9:1038. doi: 10.3389/fonc.2019.01038
102. Jiang H, Ning G, Wang Y, Lv W. Identification of an M6a-Related Signature as Biomarker for Hepatocellular Carcinoma Prognosis and Correlates With Sorafenib and Anti-PD-1 Immunotherapy Treatment Response. Dis Markers (2021) 2021:5576683. doi: 10.1155/2021/5576683
103. Shao XY, Dong J, Zhang H, Wu YS, Zheng L. Systematic Analyses of the Role of the Reader Protein of N (6)-Methyladenosine RNA Methylation, YTH Domain Family 2, in Liver Hepatocellular Carcinoma. Front Mol Biosci (2020) 7:577460. doi: 10.3389/fmolb.2020.577460
104. Lin Y, Yao Y, Wang Y, Wang L, Cui H. PD-L1 and Immune Infiltration of M(6)A RNA Methylation Regulators and Its miRNA Regulators in Hepatocellular Carcinoma. BioMed Res Int (2021) 2021:5516100. doi: 10.1155/2021/5516100
105. Qi LW, Jia JH, Jiang CH, Hu JM. Contributions and Prognostic Values of N6-Methyladenosine RNA Methylation Regulators in Hepatocellular Carcinoma. Front Genet (2020) 11:614566. doi: 10.3389/fgene.2020.614566
106. Du Y, Ma Y, Zhu Q, Liu T, Jiao Y, Yuan P, et al. An M6a-Related Prognostic Biomarker Associated With the Hepatocellular Carcinoma Immune Microenvironment. Front Pharmacol (2021) 12:707930. doi: 10.3389/fphar.2021.707930
107. Xu Y, He X, Deng J, Xiong L, Li Y, Zhang X, et al. Comprehensive Analysis of the Immune Infiltrates and PD-L1 of M(6)A RNA Methylation Regulators in Hepatocellular Carcinoma. Front Cell Dev Biol (2021) 9:681745. doi: 10.3389/fcell.2021.681745
108. Tang R, Zhang Y, Liang C, Xu J, Meng Q, Hua J, et al. The Role of M6a-Related Genes in the Prognosis and Immune Microenvironment of Pancreatic Adenocarcinoma. PeerJ (2020) 8:e9602. doi: 10.7717/peerj.9602
109. Gao W, Cheng L, He S, Li W, Zhou C, Zhou B, et al. Multiomics Integrative Analysis for Gene Signatures and Prognostic Values of M(6)A Regulators in Pancreatic Adenocarcinoma: A Retrospective Study in The Cancer Genome Atlas Project. Aging (Albany NY) (2020) 12(20):20587–610. doi: 10.18632/aging.103942
110. Zhou Z, Zhang J, Xu C, Yang J, Zhang Y, Liu M, et al. An Integrated Model of N6-Methyladenosine Regulators to Predict Tumor Aggressiveness and Immune Evasion in Pancreatic Cancer. EBioMedicine (2021) 65:103271. doi: 10.1016/j.ebiom.2021.103271
111. Xu F, Zhang Z, Yuan M, Zhao Y, Zhou Y, Pei H, et al. M6A Regulatory Genes Play an Important Role in the Prognosis, Progression and Immune Microenvironment of Pancreatic Adenocarcinoma. Cancer Invest (2021) 39(1):39–54. doi: 10.1080/07357907.2020.1834576
112. Wang L, Zhang S, Li H, Xu Y, Wu Q, Shen J, et al. Quantification of M6a RNA Methylation Modulators Pattern was a Potential Biomarker for Prognosis and Associated With Tumor Immune Microenvironment of Pancreatic Adenocarcinoma. BMC Cancer (2021) 21(1):876. doi: 10.1186/s12885-021-08550-9
113. Xu F, Huang X, Li Y, Chen Y, Lin L. M(6)A-Related lncRNAs Are Potential Biomarkers for Predicting Prognoses and Immune Responses in Patients With LUAD. Mol Ther Nucleic Acids (2021) 24:780–91. doi: 10.1016/j.omtn.2021.04.003
114. Zhou H, Zheng M, Shi M, Wang J, Huang Z, Zhang H, et al. Characteristic of Molecular Subtypes in Lung Adenocarcinoma Based on M6a RNA Methylation Modification and Immune Microenvironment. BMC Cancer (2021) 21(1):938. doi: 10.1186/s12885-021-08655-1
115. Xu F, Chen JX, Yang XB, Hong XB, Li ZX, Lin L, et al. Analysis of Lung Adenocarcinoma Subtypes Based on Immune Signatures Identifies Clinical Implications for Cancer Therapy. Mol Ther Oncolytics (2020) 17:241–9. doi: 10.1016/j.omto.2020.03.021
116. Tsuchiya K, Yoshimura K, Inoue Y, Iwashita Y, Yamada H, Kawase A, et al. YTHDF1 and YTHDF2 Are Associated With Better Patient Survival and an Inflamed Tumor-Immune Microenvironment in Non-Small-Cell Lung Cancer. Oncoimmunology (2021) 10(1):1962656. doi: 10.1080/2162402X.2021.1962656
117. Zhao J, Lu L. Interplay Between RNA Methylation EraserFTO and Writer METTL3 in Renal Clear Cell Carcinoma Patient Survival. Recent Pat Anticancer Drug Discovery (2021) 16(3):363–76. doi: 10.2174/1574892816666210204125155
118. Zhong J, Liu Z, Cai C, Duan X, Deng T, Zeng G. M(6)A Modification Patterns and Tumor Immune Landscape in Clear Cell Renal Carcinoma. J Immunother Cancer (2021) 9(2):e001646. doi: 10.1136/jitc-2020-001646
119. Xu W, Tian X, Liu W, Anwaier A, Su J, Zhu W, et al. M(6)A Regulator-Mediated Methylation Modification Model Predicts Prognosis, Tumor Microenvironment Characterizations and Response to Immunotherapies of Clear Cell Renal Cell Carcinoma. Front Oncol (2021) 11:709579. doi: 10.3389/fonc.2021.709579
120. Chen L, Hu B, Song X, Wang L, Ju M, Li Z, et al. M(6)A RNA Methylation Regulators Impact Prognosis and Tumor Microenvironment in Renal Papillary Cell Carcinoma. Front Oncol (2021) 11:598017. doi: 10.3389/fonc.2021.598017
121. Wang X, Xie H, Ying Y, Chen D, Li J. Roles of N(6) -Methyladenosine (M(6) A) RNA Modifications in Urological Cancers. J Cell Mol Med (2020) 24(18):10302–10. doi: 10.1111/jcmm.15750
122. Asada K, Bolatkan A, Takasawa K, Komatsu M, Kaneko S, Hamamoto R. Critical Roles of N(6)-Methyladenosine (M(6)A) in Cancer and Virus Infection. Biomolecules (2020) 10(7):1071. doi: 10.3390/biom10071071
123. Lin G, Zhang J, Wu Y, Zhu S, Li G. Signatures and Prognostic Values of N6-Methyladenosine (M6a) - Related Immune Genes in Bladder Cancer. Bioengineered (2021) 12(1):2649–63. doi: 10.1080/21655979.2021.1937910
124. Li Z, Li Y, Zhong W, Huang P. M6a-Related lncRNA to Develop Prognostic Signature and Predict the Immune Landscape in Bladder Cancer. J Oncol (2021) 2021:7488188. doi: 10.1155/2021/7488188
125. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell (2019) 76(1):96–109.e9. doi: 10.1016/j.molcel.2019.07.016
126. Kumar S, Nagpal R, Kumar A, Ashraf MU, Bae YS. Immunotherapeutic Potential of M6a-Modifiers and MicroRNAs in Controlling Acute Myeloid Leukaemia. Biomedicines (2021) 9(6):690. doi: 10.3390/biomedicines9060690
127. Ianniello Z, Fatica A. N6-Methyladenosine Role in Acute Myeloid Leukaemia. Int J Mol Sci (2018) 19(8):2345. doi: 10.3390/ijms19082345
128. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell (2020) 38(1):79–96 e11. doi: 10.1016/j.ccell.2020.04.017
129. Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, et al. M(6)A mRNA Demethylase FTO Regulates Melanoma Tumorigenicity and Response to Anti-PD-1 Blockade. Nat Commun (2019) 10(1):2782. doi: 10.1038/s41467-019-10669-0
130. Luo P, Pang W, Wang Y, Liu M, Zhou S, Liu S, et al. WeChat as a Platform for Problem-Based Learning Among Hematological Postgraduates: Feasibility and Acceptability Study. J Med Internet Res (2021) 23(5):e16463. doi: 10.2196/16463
131. Gu J, Bi F. Significance of N6-Methyladenosine RNA Methylation Regulators in Immune Infiltrates of Ovarian Cancer. Front Genet (2021) 12:671179. doi: 10.3389/fgene.2021.671179
132. Ma J, Yang D, Ma XX. Immune Infiltration-Related N6-Methyladenosine RNA Methylation Regulators Influence the Malignancy and Prognosis of Endometrial Cancer. Aging (Albany NY) (2021) 13(12):16287–315. doi: 10.18632/aging.203157
133. Zhao Y, Sun H, Zheng J, Shao C. Analysis of RNA M(6)A Methylation Regulators and Tumour Immune Cell Infiltration Characterization in Prostate Cancer. Artif Cells Nanomed Biotechnol (2021) 49(1):407–35. doi: 10.1080/21691401.2021.1912759
134. Wang T, Kong S, Tao M, Ju S. The Potential Role of RNA N6-Methyladenosine in Cancer Progression. Mol Cancer (2020) 19(1):88. doi: 10.1186/s12943-020-01204-7
135. Lin Y, Wang S, Liu S, Lv S, Wang H, Li F. Identification and Verification of Molecular Subtypes With Enhanced Immune Infiltration Based on M6a Regulators in Cutaneous Melanoma. BioMed Res Int (2021) 2021:2769689. doi: 10.1155/2021/2769689
136. Wu XR, Chen Z, Liu Y, Chen ZZ, Tang F, Chen ZZ, et al. Prognostic Signature and Immune Efficacy of M(1) A-, M(5) C- and M(6) A-Related Regulators in Cutaneous Melanoma. J Cell Mol Med (2021) 25(17):8405–18. doi: 10.1111/jcmm.16800
137. Mehdi A, Rabbani SA. Role of Methylation in Pro- and Anti-Cancer Immunity. Cancers (Basel) (2021) 13(3):545. doi: 10.3390/cancers13030545
138. Yi L, Wu G, Guo L, Zou X, Huang P. Comprehensive Analysis of the PD-L1 and Immune Infiltrates of M(6)A RNA Methylation Regulators in Head and Neck Squamous Cell Carcinoma. Mol Ther Nucleic Acids (2020) 21:299–314. doi: 10.1016/j.omtn.2020.06.001
139. Li Y, Zheng JN, Wang EH, Gong CJ, Lan KF, Ding X. The M6a Reader Protein YTHDC2 Is a Potential Biomarker and Associated With Immune Infiltration in Head and Neck Squamous Cell Carcinoma. PeerJ (2020) 8:e10385. doi: 10.7717/peerj.10385
140. Feng ZY, Gao HY, Feng TD. Immune Infiltrates of M(6)A RNA Methylation-Related lncRNAs and Identification of PD-L1 in Patients With Primary Head and Neck Squamous Cell Carcinoma. Front Cell Dev Biol (2021) 9:672248. doi: 10.3389/fcell.2021.672248
141. Lu S, Yu Z, Xiao Z, Zhang Y. Gene Signatures and Prognostic Values of M(6)A Genes in Nasopharyngeal Carcinoma. Front Oncol (2020) 10:875. doi: 10.3389/fonc.2020.00875
142. Saeed MEM, Mertens R, Handgretinger R, Efferth T. Identification of Fatal Outcome in a Childhood Nasopharyngeal Carcinoma Patient by Protein Expression Profiling. Int J Oncol (2018) 53(4):1721–31. doi: 10.3892/ijo.2018.4491
143. Minichsdorfer C, Oberndorfer F, Krall C, Kornek G, Mullauer L, Wagner C, et al. PD-L1 Expression on Tumor Cells Is Associated With a Poor Outcome in a Cohort of Caucasian Nasopharyngeal Carcinoma Patients. Front Oncol (2019) 9:1334. doi: 10.3389/fonc.2019.01334
144. Larbcharoensub N, Mahaprom K, Jiarpinitnun C, Trachu N, Tubthong N, Pattaranutaporn P, et al. Characterization of PD-L1 and PD-1 Expression and CD8+ Tumor-Infiltrating Lymphocyte in Epstein-Barr Virus-Associated Nasopharyngeal Carcinoma. Am J Clin Oncol (2018) 41(12):1204–10. doi: 10.1097/COC.0000000000000449
145. Jin Y, Wang Z, He D, Zhu Y, Hu X, Gong L, et al. Analysis of M6a-Related Signatures in the Tumor Immune Microenvironment and Identification of Clinical Prognostic Regulators in Adrenocortical Carcinoma. Front Immunol (2021) 12:637933. doi: 10.3389/fimmu.2021.637933
146. Liu H, Qin G, Ji Y, Wang X, Bao H, Guan X, et al. Potential Role of M6a RNA Methylation Regulators in Osteosarcoma and its Clinical Prognostic Value. J Orthop Surg Res (2021) 16(1):294. doi: 10.1186/s13018-021-02422-5
147. Li J, Rao B, Yang J, Liu L, Huang M, Liu X, et al. Dysregulated M6a-Related Regulators Are Associated With Tumor Metastasis and Poor Prognosis in Osteosarcoma. Front Oncol (2020) 10:769. doi: 10.3389/fonc.2020.00769
148. Jiang Y, Xie J, Han Z, Liu W, Xi S, Huang L, et al. Immunomarker Support Vector Machine Classifier for Prediction of Gastric Cancer Survival and Adjuvant Chemotherapeutic Benefit. Clin Cancer Res (2018) 24(22):5574–84. doi: 10.1158/1078-0432.ccr-18-0848
149. Jiang Y, Xie J, Huang W, Chen H, Xi S, Han Z, et al. Tumor Immune Microenvironment and Chemosensitivity Signature for Predicting Response to Chemotherapy in Gastric Cancer. Cancer Immunol Res (2019) 7(12):2065–73. doi: 10.1158/2326-6066.cir-19-0311
150. Li TJ, Jiang YM, Hu YF, Huang L, Yu J, Zhao LY, et al. Interleukin-17-Producing Neutrophils Link Inflammatory Stimuli to Disease Progression by Promoting Angiogenesis in Gastric Cancer. Clin Cancer Res (2017) 23(6):1575–85. doi: 10.1158/1078-0432.CCR-16-0617
Keywords: N6-methyladenosine methylation, m6A, tumor immunity, immunotherapy, tumor immune microenvironment
Citation: Guo L, Yang H, Zhou C, Shi Y, Huang L and Zhang J (2021) N6-Methyladenosine RNA Modification in the Tumor Immune Microenvironment: Novel Implications for Immunotherapy. Front. Immunol. 12:773570. doi: 10.3389/fimmu.2021.773570
Received: 10 September 2021; Accepted: 25 November 2021;
Published: 09 December 2021.
Edited by:
Guohui Wan, Sun Yat-sen University, ChinaReviewed by:
Gang Xiao, Zhejiang University, ChinaMing Cui, Peking Union Medical College Hospital (CAMS), China
Chao Jack Wang, Purdue University Indianapolis, United States
Copyright © 2021 Guo, Yang, Zhou, Shi, Huang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Shi, sy_rjh@aliyun.com; Lei Huang, lei.huang@alumni.dkfz.de
†ORCID: Lei Huang, orcid.org/0000-0002-4225-9200
 Liting Guo
Liting Guo Yan Shi
Yan Shi Lei Huang
Lei Huang Jun Zhang
Jun Zhang