Characterisation of the role and regulation of Ultrabithorax in sculpting fine-scale leg morphology
- 1Department of Biological and Medical Sciences, Faculty of Health and Life Sciences, Oxford Brookes University, Oxford, United Kingdom
- 2Centre for Functional Genomics, Department of Biological and Medical Sciences, Faculty of Health and Life Sciences, Oxford Brookes University, Oxford, United Kingdom
- 3Department of Biosciences, Durham University, Durham, United Kingdom
Hox genes are expressed during embryogenesis and determine the regional identity of animal bodies along the antero-posterior axis. However, they also function post-embryonically to sculpt fine-scale morphology. To better understand how Hox genes are integrated into post-embryonic gene regulatory networks, we further analysed the role and regulation of Ultrabithorax (Ubx) during leg development in Drosophila melanogaster. Ubx regulates several aspects of bristle and trichome patterning on the femurs of the second (T2) and third (T3) leg pairs. We found that repression of trichomes in the proximal posterior region of the T2 femur by Ubx is likely mediated by activation of the expression of microRNA-92a and microRNA-92b by this Hox protein. Furthermore, we identified a novel enhancer of Ubx that recapitulates the temporal and regional activity of this gene in T2 and T3 legs. We then used transcription factor (TF) binding motif analysis in regions of accessible chromatin in T2 leg cells to predict and functionally test TFs that may regulate the Ubx leg enhancer. We also tested the role of the Ubx co-factors Homothorax (Hth) and Extradenticle (Exd) in T2 and T3 femurs. We found several TFs that may act upstream or in concert with Ubx to modulate trichome patterning along the proximo-distal axis of developing femurs and that the repression of trichomes also requires Hth and Exd. Taken together our results provide insights into how Ubx is integrated into a post-embryonic gene regulatory network to determine fine-scale leg morphology.
Introduction
The Hox genes encode an important and conserved family of transcription factors (TFs) that are expressed during embryogenesis to determine the identity of body regions along the antero-posterior (A-P) axis of animals (Lewis, 1978; McGinnis and Krumlauf, 1992; Pearson et al., 2005; Krumlauf, 2018). Hox genes also play more subtle but important post-embryonic roles in regulating cell identify to sculpt the fine-scale morphology of structures and organs, and consequently have been likened to “micromanagers” (Akam, 1998a; Akam, 1998b; Hombría and Lovegrove, 2003; Buffry and McGregor, 2022). Several such post-embryonic roles of Hox genes have been identified in Drosophila; for example, the specification of certain subtypes of cells in the central nervous system (Kannan et al., 2010; Estacio-Gómez et al., 2013), the regulation of the development of larval oenocytes by abdominal-A (abd-A) (Brodu et al., 2002), and the integration of regulatory information to specify differences in prothoracic (T1) leg bristle patterning among leg segments and between sexes by Sex-combs reduced (Scr) (Eksi et al., 2018).
Ultrabithorax (Ubx) specifies the identity of thoracic and abdominal segments during Drosophila embryogenesis (White and Wilcox, 1984; Akam and Martinez-Arias, 1985; White and Wilcox, 1985; Castelli-Gair and Akam, 1995). Classically, this Hox gene represses wing identity and promotes haltere formation on the third thoracic (T3) segment through the direct regulation of potentially hundreds of genes (White and Wilcox, 1984; White and Akam, 1985; White and Wilcox, 1985; Weatherbee et al., 1998; Pavlopoulos and Akam, 2011; Diaz-de-la-Loza et al., 2020). Ubx also distinguishes the size and morphology of halteres at a more fine-scale level, in part through the autoregulation of differences in the expression levels between proximal and distal cells (Roch and Akam, 2000; Delker et al., 2019). In these appendages Ubx also influences chromatin accessibility through cell type specific interactions with co-factors and can thereby act as a repressor as well as an activator (Loker et al., 2021). During mesothoracic (T2) and T3 leg development, Ubx is expressed along the proximo-distal axis of pupal femurs, with the highest concentration in proximal-posterior and dorsal-anterior cells (Stern, 1998; Davis et al., 2007). This expression of Ubx regulates the patterning of trichomes and bristles on the T2 and T3 femurs in a concentration dependent manner (Morata and Kerridge, 1981; Kerridge and Morata, 1982; Struhl, 1982; Casanova et al., 1985; Stern, 1998; Stern, 2003; Davis et al., 2007; Shroff et al., 2007; Schubiger et al., 2012). Therefore, in addition to determining segmental identity, Ubx subsequently contributes to sculpting the finer-scale morphology of several appendages.
Despite these insights into Hox gene function, we still do not fully understand how they are integrated into post-embryonic gene regulatory networks (GRNs). One approach to address this is to study the regulation of Hox genes by identifying the enhancers that are responsible for their post-embryonic expression. Indeed, several enhancers and other cis-regulatory elements of Ubx have already been identified, including those corresponding to classic mutations with phenotypic effects, such as anterobithorax (abx), postbithorax (pbx) and bithorax (bx), and we are beginning to understand how they integrate information to precisely regulate the differential expression of this Hox gene to control fine-scale morphology (Bender et al., 1983; Peifer and Bender, 1986; Little et al., 1990; Simon et al., 1990; Irvine et al., 1991; Müller and Bienz, 1991; Irvine et al., 1993; Pirrotta et al., 1995; Maeda and Karch, 2006; Magbanua et al., 2015; Delker et al., 2019). However, it is clear that not all Ubx enhancers have been identified and we still have much to learn about the complex regulation of this crucial gene (Davis et al., 2007; Magbanua et al., 2015; Delker et al., 2019).
Enhancers can be challenging to identify because currently there is no consensus of what genomic features mark these regions (Buffry et al., 2016; Halfon, 2019). Furthermore, although the regulatory genome can now more readily be studied with new tools such as ATAC-seq, C technologies and CRISPR/Cas9, we still do not fully understand the regulatory logic underlying enhancer function (Buffry et al., 2016; Halfon, 2019; Bolt and Duboule, 2020; Jindal and Farley, 2021). Given their importance in development, disease and evolution, it is crucial that we continue to identify and study individual enhancers in detail, to better our general understanding of cis-regulatory regions and GRNs.
The development and patterning of trichomes among Drosophila species has proven an excellent model to study GRNs and their evolution (Stern and Frankel, 2013; Arif et al., 2015; Kittelmann et al., 2021). Trichomes are short, non-sensory, actin protrusions that are found on insect bodies throughout all stages of life (Arif et al., 2015). They are thought to be involved in processes such as aerodynamics, thermal regulation and larval locomotion (Balmert et al., 2011; Ditsche-Kuru et al., 2011). The larval cuticle of Drosophila displays a distinct pattern of trichomes and the underlying GRN is understood in great detail (Delon et al., 2003; Chanut-Delalande et al., 2006; Menoret et al., 2013). In brief, the gene shavenbaby (svb) appears to integrate information from upstream factors, including Ubx, and directs expression of downstream effector genes that determine the formation of the trichomes themselves (Delon et al., 2003; Chanut-Delalande et al., 2006; Menoret et al., 2013; Crocker et al., 2015; Preger-Ben Noon et al., 2016). Moreover, the convergent evolution of larval trichome patterns in different Drosophila lineages is caused by changes in enhancers of svb (Sucena and Stern, 2000; Sucena et al., 2003; McGregor et al., 2007; Frankel et al., 2011; Frankel et al., 2012; Stern and Frankel, 2013; Crocker et al., 2015; Preger-Ben Noon et al., 2016).
The T2 legs of D. melanogaster display a trichome pattern with a patch of cuticle on the proximal posterior of the femur that is free from trichomes, known as the “naked valley” (NV) (Stern, 1998; Arif et al., 2013) (Figure 1A). We previously studied the GRN underlying leg trichome patterning and found that it differs in topology with respect to the larval trichome GRN (Kittelmann et al., 2018). In particular, in the developing T2 legs, the Svb-dependent activation of trichomes is blocked by microRNA-92a (miR-92a)-mediated repression of Svb target genes to generate the NV (Arif et al., 2013; Kittelmann et al., 2018). Furthermore, in contrast to its activation of the larval trichomes, Ubx represses leg trichomes perhaps via miR-92a (Stern, 1998; Arif et al., 2013; Crocker et al., 2015).
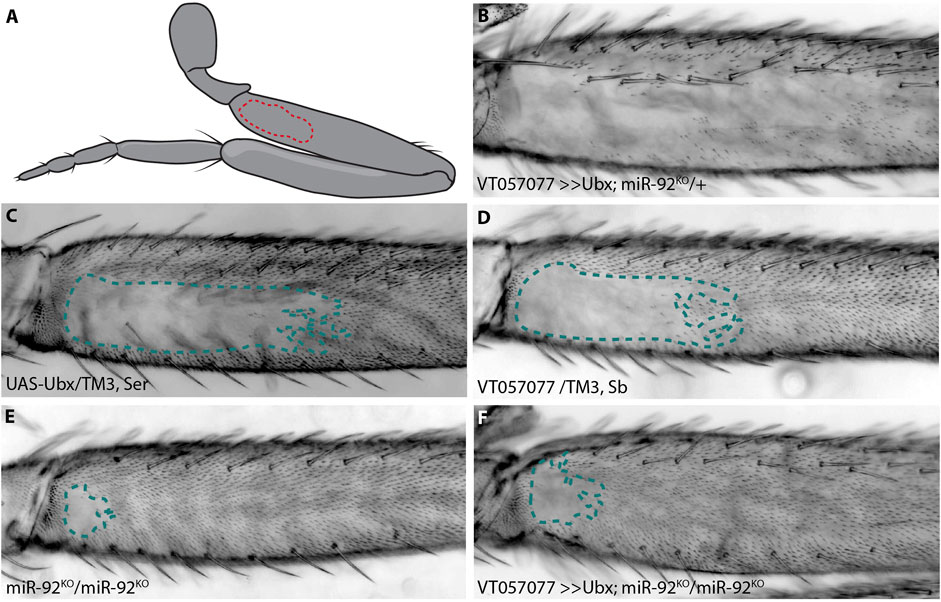
FIGURE 1. Ubx requires miR-92 genes to repress trichomes. The naked valley is a region of trichome-free cuticle on the posterior side of the proximal femur (outlined by dotted lines) (A). Over-expression of Ubx inhibits trichome formation on most of the T2 femur (B) while the UAS-Ubx line (C) and the GAL4 driver line VT057077 (D) both have large naked valleys. Naked cuticle is almost absent in miR-92 loss of function T2 legs (E). Trichome inhibition by Ubx over-expression is restricted to the most proximal region of the femur in miR-92KO (F).
The size of the NV varies within and between species and these differences are associated with changes in the expression of miR-92a (Arif et al., 2013) and Ubx (Stern, 1998), respectively. Ubx is expressed in D. melanogaster T2 legs in the region of the NV, but not in the T2 legs of D. virilis, which has no NV (Stern, 1998). Moreover, it has been shown that Ubx contributes to differences in NV size between D. melanogaster and D. simulans (Stern, 1998). It was postulated that the evolution of Ubx expression in T2 legs is attributable to the presence of a T2 leg-specific enhancer of Ubx (Davis et al., 2007). However, no cis-regulatory sequences that could drive expression of Ubx in T2 were identified.
Here we further characterise how Ubx is wired into the GRN for leg trichome patterning. We show that repression of trichomes by Ubx is likely dependent on activation of miR-92a and potentially its closely linked paralogue miR-92b by this Hox gene. We also identified a novel enhancer of Ubx that drives expression along the proximo-distal axis of T2 and T3 femurs during trichome patterning. Functional analysis of TFs predicted to bind to this Ubx leg enhancer revealed that several activate or repress leg trichomes and that repression of trichomes by Ubx is dependent on the co-factors Extradenticle (Exd) and Homothorax (Hth). Taken together our results provide new insights into the role and regulation of Ubx during post-embryonic development and in sculpting fine-scale adult morphology.
Materials and methods
Fly stocks and genetics
All stocks used were kept on standard yeast extract-sucrose medium at 25°C. Reporter lines VT42732, VT42733, and VT42734 were obtained from the Vienna Drosophila Resource Centre (VDRC). Lines GMR31F12, GMR32BO3 and GMR31E11 were obtained from the FlyLight enhancer collection (Jenett et al., 2012). To test the activity of all enhancer lines, they were crossed to a UAS-stingerGFP and/or UAS-shavenoid (sha)
Cloning
Fragments UbxP1, e33.A, e33.B and e33.C were PCR-amplified from genomic DNA (D. melanogaster, Oregon R). UbxP1 was cloned directly into the S3aG expression vector (a gift from Thomas Williams, Addgene plasmid #31171). Fragments, e33.A, e33.B and e33.C, were initially inserted into the TOPO/D vector (Invitrogen). LR gateway cloning was then used to subclone the fragments into the pBPGUw plasmid upstream of GAL4 (a gift from Gerald Rubin, Addgene plasmid #17575). The resulting constructs were inserted into landing site 86Fb using phiC31 mediated germline transformation by either BestGene Inc. or the Cambridge injection facility. Genomic coordinates of fragments and primer sequences can be found in Supplementary Material S2.
GFP and NV analysis
To assay expression in pupae, white prepupae from GAL4 lines crossed to UAS-stingerGFP were collected and aged to between 20 and 28 h after puparium formation (hAPF), the window when T2 trichome patterning is regulated by Ubx (Stern, 1998). UbxP1 flies were analysed without crossing to UAS-GFP as they were constructed in such a way to allow direct GFP expression. GFP expressing whole pupae were imaged on a Zeiss Axiozoom stereoscope. For the dissection of pupal legs, the pupal case was removed and the pupae were covered in 4% formaldehyde for 10 min, a tungsten needle was then used to create small openings in the head and abdomen. Fresh 4% formaldehyde was flushed over the pupae and left for another 10 min. T2 and T3 pupal legs were then dissected with tungsten needles and mounted in 80% glycerol. Mounted legs were immediately imaged on a Zeiss 800 confocal. For the analysis of expression patterns in T2 and T3 leg discs, 3rd instar larvae were dissected and reporter expression visualised with anti-GFP (Thermofisher) (1:600) and goat anti-chicken 488 (1:600) according to standard protocols. Discs were also stained with DAPI and mounted in 80% glycerol and imaged on a Zeiss 800 confocal. For the analysis of trichome patterns, T2 and T3 legs were dissected from adults and mounted in Hoyer’s medium/lactic acid (1:1) and imaged under a Zeiss Axioplan microscope with a ProgRes MF cool camera (Jenaoptik). The size of the NV was measured (n = at least 10) using Fiji software (Schindelin et al., 2012) and statistical analysis was performed in R-Studio version 1.2.1335 (RCoreTeam, 2022). We expect the NV of the progeny to be an intermediate size between the two parental lines, if this was not the case further statistics were carried out. Data were checked for normality using Shapiro-Wilk, followed by either an ANOVA or Kruskal–Wallis test. To check significance between each group either Tukey’s post-hoc test or Dunn’s test was performed, p-values were adjusted using Bonferroni correction to avoid multiple testing errors. For SEM imaging, legs were dissected from adult flies and stored in fresh 100% ethanol. Legs were then critically point dried using automatic mode of a Tousimis 931.GL Critical Point Dryer, mounted on SEM stubs with carbon tabs, sputter-coated with a 15 nm thick gold coat and imaged in a Hitachi S-3400N at 5 kV with secondary electrons.
Identification and functional testing of candidate TFs
To identify potential TFs that bind to the Ubx leg enhancer, the JASPAR TF database was utilised (Fornes et al., 2020) with a relative profile threshold of 85% similarity. Each position weight matrix (PWM) for a given TF is scored based on the similarity between the pattern of nucleotides in the motif and DNA sequences of the same length, the higher the score, the better the similarity. By setting the relative profile threshold to 85%, only PWMs with a score greater than, or equal to 85% will be provided. The resulting factors were compared to the RNA-seq data for T2 legs (GEO accession number GSE113240) (Kittelmann et al., 2018), and genes encoding TFs with an expression level of over 1 fragment per kb per million (FKPM) were scored as expressed. To further filter TFs, only those with predicted binding sites in regions of accessible chromatin from T2 leg ATAC-seq profiling data (Supplementary Material S3) (GEO accession number GSE113240) (Kittelmann et al., 2018) were selected. To assay whether the identified TFs have any role in trichome development on the T2 and T3 legs, RNAi lines for selected genes were crossed to VT33-GAL4 enhancer and the resulting trichome pattern was measured and compared to parental control lines (Supplementary Material S4).
Results
Ubx repression of T2 leg trichomes requires miR-92 genes
In addition to its well characterised role in T3 leg development, it was previously found that Ubx represses the formation of trichomes on T2 femurs in a dose sensitive manner from proximal to distal (Stern, 1998). We corroborated this finding by over-expressing Ubx in T2 legs, which resulted in loss of all proximal and most of the distal trichomes on posterior T2 femurs, including those dorsal and ventral of the NV (Figures 1A–D). We hypothesised previously that this repressive effect is mediated through a microRNA, miR-92a. This microRNA gene and its paralogue miR-92b are located in an intron and the 3′UTR of the protein-coding gene, jing interacting regulatory 1 (jigr1), respectively. They also have the same seed sequence and, therefore, likely the same target genes, and here we refer to both paralogues collectively as miR-92. As we showed previously, over-expression of miR-92a also represses T2 trichomes and, reciprocally, loss of both microRNAs results in a very small NV (Arif et al., 2013; Kittelmann et al., 2018) (Figure 1E). This suggests that Ubx acts upstream of miR-92 to inhibit trichome formation. In order to test this, we over-expressed Ubx in flies homozygous for a loss of function of miR-92 (Yuva-Aydemir et al., 2015). We found that Ubx is unable to repress trichomes in the absence of these microRNAs (Figure 1F).
We also tested the effects of Ubx over-expression and miR-92 loss of function on T3 leg trichomes (Supplementary Figure S1). Without miR-92, trichomes develop in normally naked regions of the posterior T3 femur, albeit in a patchy pattern (Supplementary Figure S1). This is also the case when Ubx is over-expressed in the absence of miR-92, indicating again that Ubx requires miR-92 to repress trichome development on the posterior of T3 femurs. Note that Ubx over-expression never interferes with the formation of anterior trichomes on T2 or T3 femurs (Supplementary Figure S1). Taken together, our findings suggest that Ubx represses trichomes on posterior femurs by directly or indirectly activating miR-92 expression, which in turn inhibits the expression of Svb target genes including shavenoid (sha) (Schertel et al., 2012; Arif et al., 2013; Kittelmann et al., 2018). To better understand how Ubx is integrated into the leg trichome GRN, we next attempted to identify cis-regulatory elements that regulate expression of this Hox gene in T2 and T3 legs.
Several regions of the Ubx locus with open chromatin drive expression in Drosophila pupal legs
To identify the previously predicted leg Ubx enhancer, Davis et al. (2007) assayed available regulatory mutations of the Ubx locus and generated new deficiencies. This allowed them to rule out approximately 100 kb in and around the Ubx locus as containing the T2 leg enhancer. They then assayed a further 30 kb using reporter constructs (Figure 2A). In total they investigated over 95% of the Ubx locus but were unable to identify a region with T2 leg specific activity.
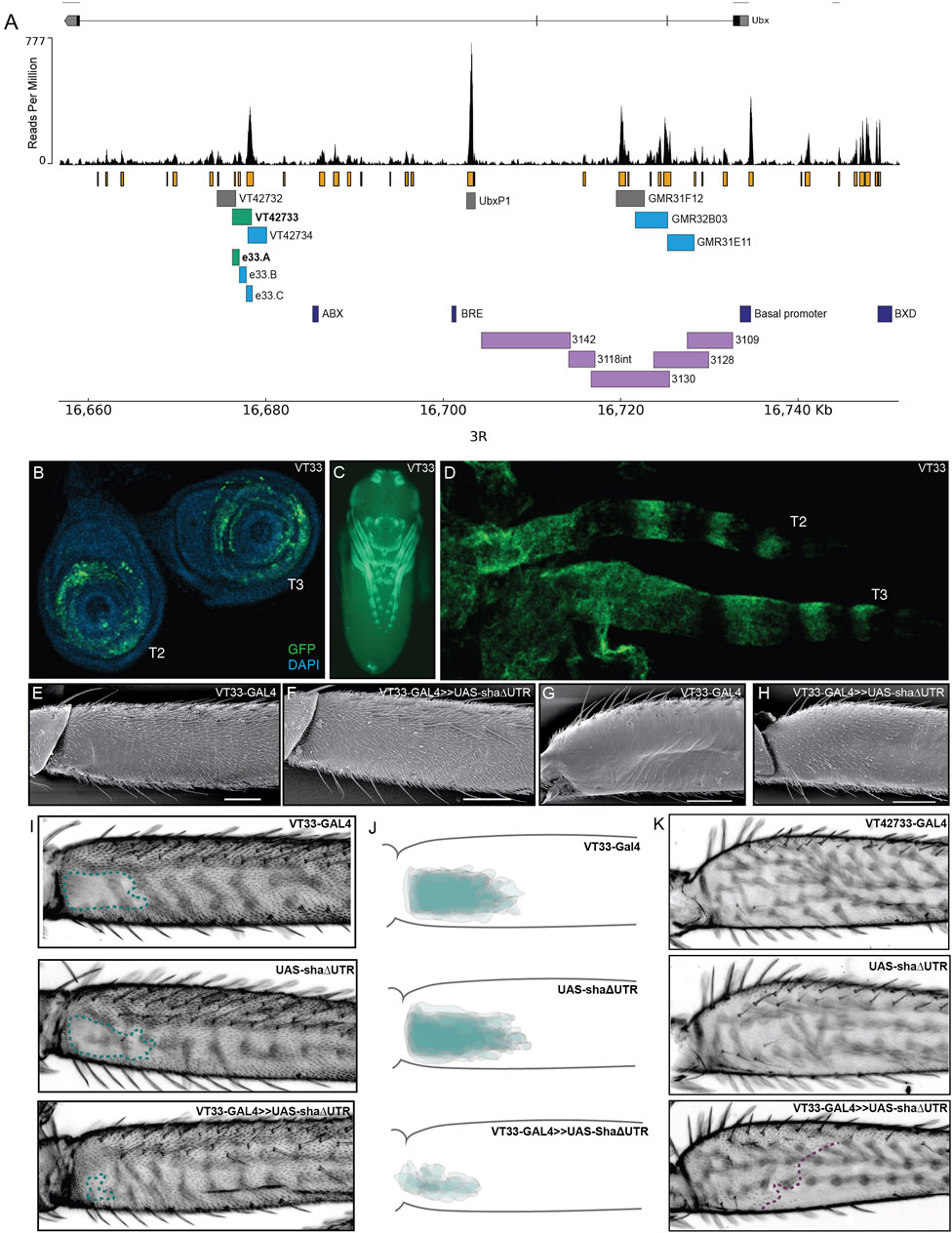
FIGURE 2. Testing regions of accessible chromatin at the Ubx locus for enhancer activity. (A) The Ubx locus in Drosophila melanogaster on chromosome arm 3R with the ATAC-seq profile for Ubx in pupal legs below. In orange are peaks of open chromatin and underneath are the locations of tested or known regulatory elements. In green are the lines tested in this study that affect trichome patterning on T2 and T3 legs. In blue are the lines which express GFP in pupal legs but do not have a functional effect on trichome patterning. In grey are the lines which do not drive expression in pupal legs. In dark blue are a subset of the characterised regulatory elements of Ubx. For a complete list of all characterised elements according to RedFly see Supplementary Material S2. In purple are the reporter constructs that were tested by Davis et al. (2007) that did not contain the leg specific enhancer of Ubx. (B) UAS-GFP expression driven by VT42733-GAL4 (referred to as VT33-GAL4) in T2 and T3 leg imaginal discs from 3rd instar larvae is seen in the femur ring of the disc but also other leg segments. (C) Expression driven by VT33-GAL4 in whole pupae at 24 hAPF. Expression is present in several different tissues including the pupal legs, antennae, mouthparts, eyes, and genitalia. (D) In T2 and T3 pupal legs from flies at 24 hAPF, the expression driven by VT33-GAL4 is observed in the developing femur and also in a striped pattern more distally in the pupal leg. (E) Scanning electron micrograph (SEM) showing the NV of a wild-type T2 proximal femur. (F) SEM of a T2 femur when the VT33 enhancer is crossed to the trichome activating line, UAS-shaΔUTR. Most of the naked cuticle is now filled with trichomes. (G) SEM of a wild-type T3 femur shows that the posterior region of the leg is largely free of trichomes. (H) When VT33-GAL4 is crossed to UAS-shaΔUTR, there is ectopic development of trichomes on the proximal part of the T3 femur. (I) Light-microscope images of adult T2 legs from progeny of the VT33-GAL4 crossed to UAS-shaΔUTR. The parental controls are shown on top and the progeny of the cross beneath. In each case the naked valley is outlined with a green dashed line. (J) Visual representation of VT33-GAL4 crossed to UAS-shaΔUTR and controls (n = 10). The T2 femurs of approximately 10 individuals were aligned and the NV was outlined and overlaid to give a visual representation of the shape and size of the NV within a treatment versus controls. There is a dramatic decrease in the size of the NV in all individuals. (K) Images of the T3 femur when VT33 is crossed to the trichome activating line versus the parental control lines. The purple dashed line indicates the extent of proximal ectopic trichomes.
To follow up the work of Davis et al. (2007), we used ATAC-seq data that we generated previously to identify regions of accessible chromatin in T2 leg cells in the developmental window when the trichome pattern is determined (Kittelmann et al., 2018) (Figure 2A). We found that the Ubx locus contains several regions of accessible chromatin in T2 pupal cells, corresponding to known enhancers or promoters as well as putative new cis-regulatory elements (Figure 2A; Supplementary Material S2).
We then took advantage of existing reporter lines (Pfeiffer et al., 2008; Jenett et al., 2012), to assay regions of open chromatin in the introns of Ubx for enhancer activity, and specifically to test if any could drive expression in developing T2 legs (Figure 2A). We selected three lines from the GMR database that overlap with lines tested by Davis et al. (2007), but did not show enhancer activity in T2 legs (Figure 2A). We also assayed three lines from the VT-GAL4 database (VDRC) corresponding to several peaks of open chromatin but not overlapping with any known regulatory elements of Ubx (RedFly: Rivera et al., 2019) (Supplementary Material S2). Finally, we tested the UbxP1 peak of accessible chromatin, which corresponds to a previously characterised variably occupied CTCF site (Magbanua et al., 2015). This region was not covered by Davis et al. (2007) and therefore was not previously tested for enhancer activity in legs. Four of the seven regions tested were able to drive reporter gene expression in developing legs at 24 hAPF, although all of them appeared to be quite promiscuous and had activity in other pupal tissues (Figures 2A, C; Supplementary Figure S2).
We next tested whether regions VT42733, VT427734, GMR32B03 and GMR31E11 (Figure 2A), which drive GFP expression in pupal legs, could also influence the trichome pattern on the femurs of T2 and T3 legs, which would further indicate that they are active in leg epidermal cells at the time of trichome patterning. To do this we crossed the driver lines to UAS-shaΔUTR, which overrides trichome repression by miR-92a and leads to the formation of trichomes on normally naked cuticle (Arif et al., 2013). Therefore, in this assay, enhancer regions that are active in posterior femurs at the correct time will generate trichomes where there is normally naked cuticle. Only one of the reporter lines identified, VT42733, was able to induce the formation of trichomes in the NV, resulting in a striking decrease in the size of the patch of naked cuticle (Figures 2A, E, F, I, J). Importantly, we noticed that while VT42733 greatly reduces the size of the NV in this assay, a small patch of naked cuticle remains proximally on the ventral side of the T2 posterior femur (Figures 2F, I, J), which is consistent with Ubx-independent repression of trichomes in these cells (Davis et al., 2007). We also observed that VT42733 was able to induce the formation of trichomes proximally on the posterior and dorsal-anterior of T3 femurs, suggesting that this enhancer also contributes to T3 femur patterning (Figures 2G, H, K). The proximal dorsal-anterior activity of VT42733 in T3 femurs overlaps with the activity of abx (Davis et al., 2007). We observed that the activity of VT42733 in the T3 femur is proximally restricted and does not extend as distally as where Ubx is known to repress trichomes, which is consistent with previous data showing that pbx and potentially bx also regulate expression of this Hox gene in the posterior of T3 femurs (Davis et al., 2007).
We examined the expression driven by VT42733 in more detail in leg imaginal discs and in pupal legs (Figures 2B, D). In 3rd instar leg discs, GFP expression driven by this enhancer can clearly be seen in rings which will develop into the future T2 and T3 femurs (Figure 2B). Similar reporter expression can be seen in the pupal T2 and T3 femurs, as well as more distal segments (Figure 2D), which is consistent with the fact that this region can promote trichome formation on T2 and T3 femurs when combined with UAS-shaΔUTR (Figures 2E–K).
Taken together these results are evidence that VT42733 represents a novel Ubx leg enhancer, which regulates expression of this Hox gene in the NV region of T2 femurs as well as proximally in T3 femurs.
Delineation of the Ubx leg enhancer
VT42733 drives expression in T2 and T3 legs consistent with Ubx activity, but this enhancer is also active in other pupal tissues (Figure 2C). To further delineate the Ubx leg enhancer region, VT42733 was subdivided into three partially overlapping fragments of around 700 bp: e33.A, e33.B, and e33.C (Figure 2A). All three lines were able to drive reporter expression in developing pupae (Figure 3; Supplementary Figure S2): e33.A drives expression in leg discs, pupal legs, antennae and developing eyes (Figures 3A, B), and e33.B drives a more restricted expression pattern limited to a small patch in the pupal legs and in the head (Supplementary Figure S2). While e33.C also drives expression in the legs, its activity is predominantly in the head and thorax as well as a stripe-like pattern on the ventral side of the abdomen, which was not seen with any of the other driver lines tested (Supplementary Figure S2). Given the intriguing and varied expression patterns displayed by subdivisions of VT42733 we sought to test if any of these regions could induce trichomes in the NV.
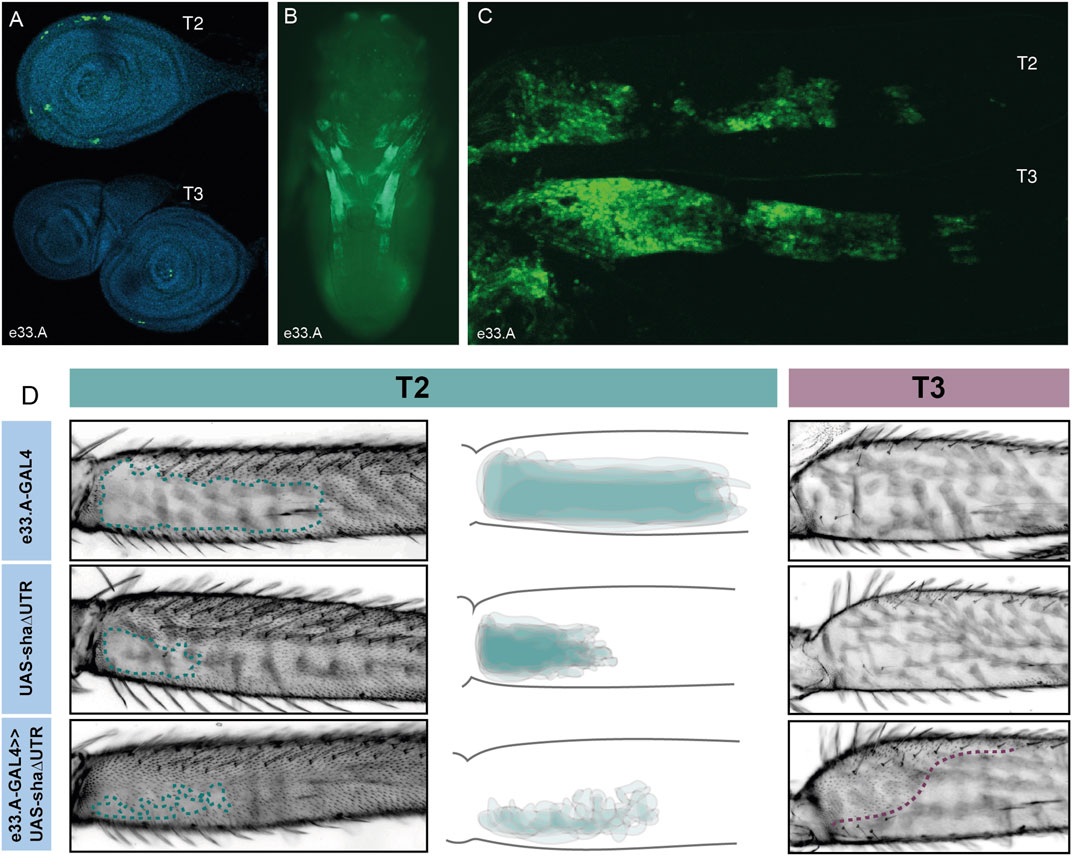
FIGURE 3. Characterisation of e33.A enhancer activity. (A) Expression driven by e33.A in T2 and T3 leg imaginal discs from 3rd instar larvae is restricted in comparison to the whole VT42733 enhancer. There is also reduced expression in T3 compared to T2. (B) In whole pupae at 24 hAPF, e33.A drives mainly in the developing legs. (C) In T2 and T3 pupal legs at 24 hAPF, there is clear expression driven by e33.A in the femur. (D) The result of crossing e33.A to the trichome activating line, UAS-shaΔUTR. On T2 legs (labelled in green), there is patchy ectopic trichome development in the NV when compared with parental controls. There is also ectopic trichome development in the proximal part of the T3 femur (outlined in purple), although this does not extend as far ventrally as with the VT42733 enhancer.
To do this they were combined with UAS-shaΔUTR. We observed that e33.A was able to drive trichomes in the T2 NV albeit in a patchy and irregular pattern compared to VT42733 (Figures 2, 3). e33.A also drove trichomes proximally in the dorsal-anterior and posterior of T3 femurs although this activity did not extend as far distally as with VT42733 (Figures 2, 3). e33.B and e33.C did not have any detectable activity in this assay (Supplementary Figure S2). This suggests at least part of the enhancer activity of VT42733 in developing T2 and T3 femurs is determined by TF binding sites (TFBS) in e33.A.
Analysis of TFs that may bind to the Ubx leg enhancer
To further characterise the Ubx leg enhancer, we carried out motif analysis to identify TFs that may bind to this region. To focus on binding sites for TFs that are expressed at the time of trichome development, we cross-referenced previously generated RNA-seq data for T2 legs (Kittelmann et al., 2018) with the JASPAR database (Fornes et al., 2020) (with the caveat that the JASPAR database does not contain an exhaustive list of all Drosophila TFs). Using a threshold of 85% similarity and focussing only on TFs expressed above a 1 FPKM threshold in T2 legs, 62 TFs were found to have predicted binding sites in the VT42733 region. We then further filtered the TFs using T2 pupal leg ATAC-seq data (Kittelmann et al., 2018) to shortlist TFs with predicted binding sites located only in the accessible chromatin of region VT42733. This resulted in a total of 55 TFs that are expressed in pupal T2 legs and predicted to bind to accessible regions in the VT42733 enhancer (Supplementary Material S3). We hypothesised that some of these TFs could be involved in the GRN underlying trichome formation.
We tested the role of 25 of these TF candidates [selected from among those predicted above after excluding those with only one predicted binding site in VT42733 (except Dichaete) as well as genes without a characterised function (“CG genes”)], and the known Ubx co-factor homothorax (hth) in T2 and T3 femur patterning by knocking down their expression using RNAi (Supplementary Materials S3, S4). To do this we used VT33-GAL4 since it expresses in the NV cells in the correct window of pupal leg development. This means that offspring of UAS-RNAi lines and VT33-GAL4 produce double-stranded RNA hairpins in NV cells, leading to cell specific silencing of genes. We found that knockdown of 8/26 genes affected the trichome pattern on the posterior of T2 femurs when normalised for femur length: arrowhead (awh), C15, Distal-less (Dll), extradenticle (exd), hth, mirror (mir), NK7.1 and ventral-veins lacking (vvl) (Figures 4, 5, 6; Supplementary Figures S3, S4; Supplementary Material S4). We note that it remains possible that more of these TFs regulate trichome patterning because some of the RNAi lines might not give detectable effects and therefore could be false negatives.
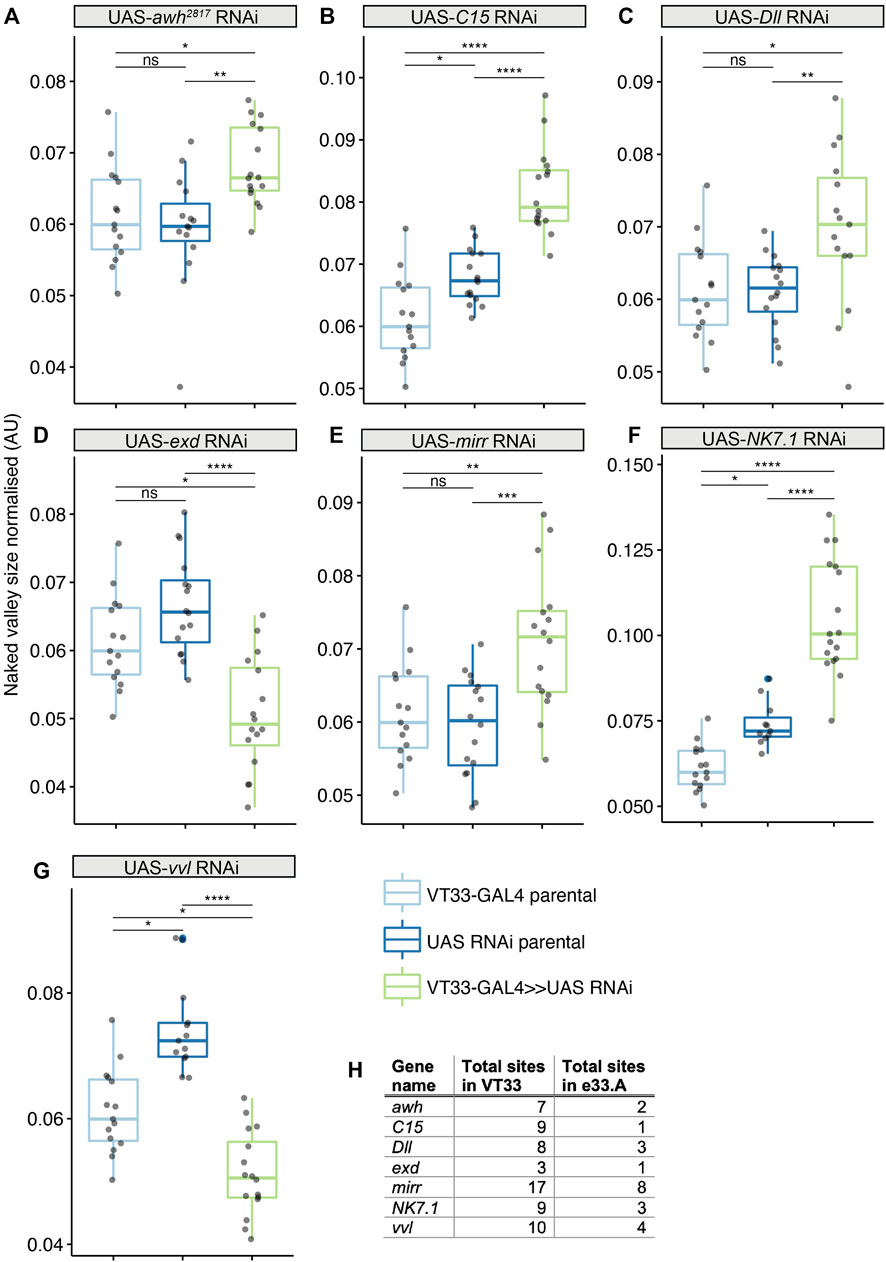
FIGURE 4. Transcription factors predicted to bind the Ubx leg enhancer that significantly affect T2 trichome patterning. Knockdown of awh (A), C15 (B), Dll (C), mirr (E) and NK7.1 (F) results in a significant increase in the size of the NV, while exd (D) and vvl (G) knockdown makes the NV smaller. In each case the progeny from the cross between the UAS-RNAi and the VT33-GAL4 (green boxes) were compared to the parental strains (blue boxes). Significance levels are shown above the pairs, p-values (p > 0.05 NS, p ≤ 0.05 *, p ≤ 0.01 **, p ≤ 0.001 ***, p ≤ 0.0001 ****) were calculated with an ANOVA or Kruskal–Wallis depending on normality. (H) Summary of the number of binding sites found by JASPAR for the seven TFs in the whole of the VT42733 (VT33) enhancer and how many of those sites are found in e33.A.
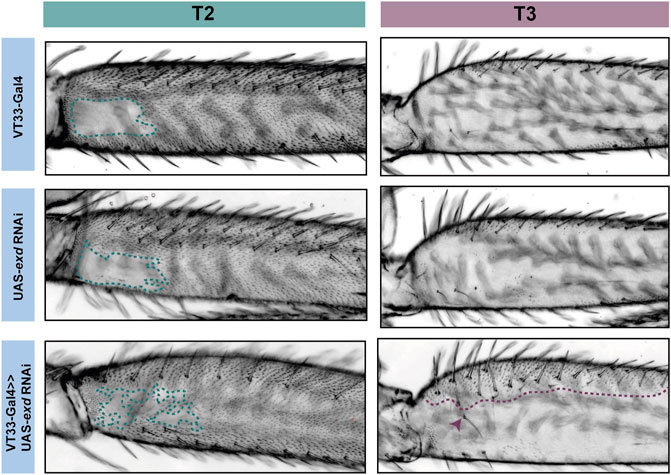
FIGURE 5. RNAi knockdown of exd results in ectopic trichome growth. exd RNAi results in patchy ectopic trichomes in the NV of T2 femurs (green). On T3 femurs (purple), exd RNAi also causes the development of ectopic trichomes, which extend about one-third from the proximal dorsal towards the ventral (purple dotted line). There is also an additional row of bristles as well as ectopic bristle growth (arrowhead).
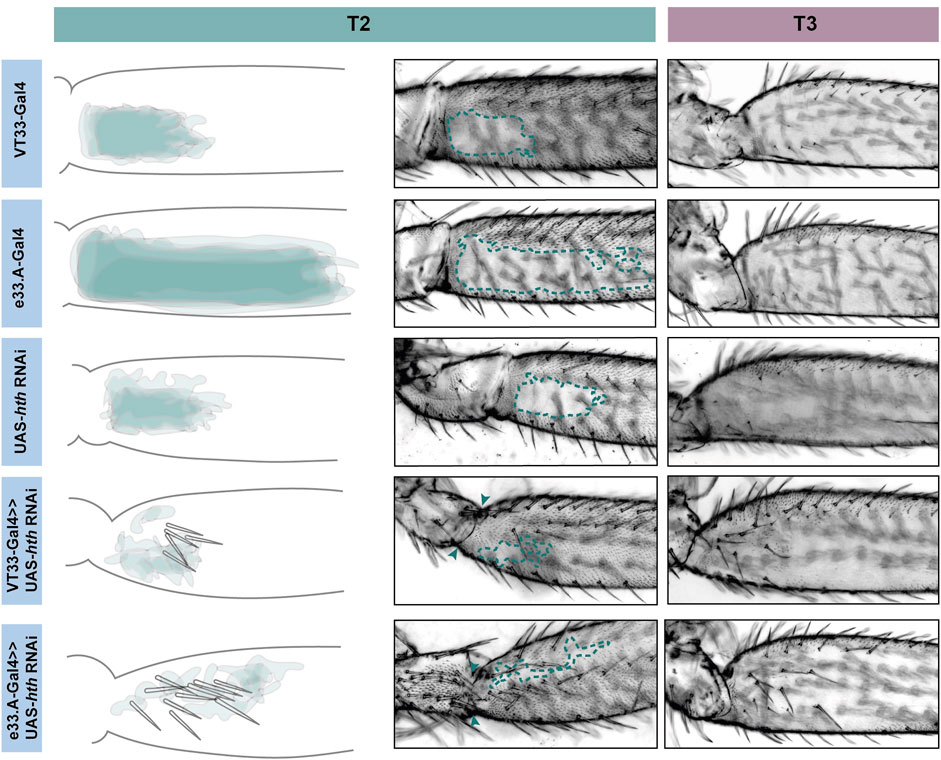
FIGURE 6. Knockdown of hth affects leg morphology and patterning. In T2 (green) knockdown of hth causes the ectopic development of trichomes and sensory bristles in the NV. There is also a change in the shape of the coxa and femur suggesting a transition to a more T1 like phenotype. This effect is more severe when the knockdown is driven with e33.A-GAL4; there is also the development of trichomes and bristles on the coxa. On T3 (purple) knockdown of hth similarly results in ectopic sensory bristles and trichomes that are restricted to the proximal of the femur.
Knockdown of exd and hth resulted in ectopic trichomes at the proximal posterior of T2 femurs (Figures 5, 6). hth knockdown had a stronger effect than exd on T2 morphology, with ectopic sensory bristles and transformation of the shape of the coxa to a more T1 like appearance (Figure 6). Surprisingly these effects were even more pronounced when using e33.A as a driver (Figure 6). On T3, knockdown of exd resulted in trichome formation in the dorsal-anterior of the femur and additional bristles along the A-P boundary (Figure 5). Knockdown of hth again had a stronger effect on T3 with additional trichomes and bristles on the dorsal-anterior and proximally on the posterior femur (Figure 6). These results are consistent with loss of Ubx function in T2 and T3 femurs (Davis et al., 2007) and suggest that Hth and Exd promote Ubx activity in T2 and T3 perhaps by acting as co-factors for this Hox gene in this context.
vvl was the only other TF tested that resulted in ectopic trichome formation in the proximal posterior of T2 femurs when knocked down (Figure 4; Supplementary Figures S3, S4). RNAi against vvl also produced ectopic trichomes in the dorsal-anterior of T3 femurs (Supplementary Figure S5). This indicates that Vvl suppresses trichome formation in T2 and T3 femurs perhaps by promoting Ubx expression or acting downstream of this Hox gene.
We found that RNAi knockdown of awh, C15, Dll, mir and NK7.1 resulted in a distal expansion of the NV on T2 femurs (Figure 4: Supplementary Figures S3, S4; Supplementary Material S4), but had no effect on T3 femur patterning (Supplementary Figure S5). This suggests that these TFs may repress Ubx activity in the distal of the posterior T2 femur or promote trichome formation in this region.
Discussion
Identification of a Ubx leg enhancer
We have found that Ubx represses trichomes on the femurs of T2 and T3 legs via mir-92 (Figure 7). We then sought to determine how this Hox gene is regulated in these appendages. We found that the 2.2 kb region VT42733 has enhancer activity in the proximal posterior of T2 and T3 femurs and proximal dorsal-anterior of T3 femurs during the correct developmental time point and consistent with Ubx functions in these legs (Figures 2E–K). Analysis of sub-fragments of VT42733 showed that a 700 bp region, e33.A, is also active in T2 and T3 femurs cells, but this activity is weaker than the full VT42733 sequence (Figure 3D) and has fewer predicted TF binding sites (Supplementary Material S3). Taken together, these results indicate that the Ubx leg enhancer is located in region VT42733 with some binding sites concentrated in region e33.A (Supplementary Material S3). Importantly, while VT42733 and e33.A are able to drive expression in the proximal femur, they are inactive in the ventral part of the posterior T2 and T3 femurs (Figures 2F, I, 3D). This is consistent with previous studies showing that while Ubx represses trichomes on the posterior T2 femurs, it is inactive in these ventral cells, and even in the absence of Ubx, this region of the cuticle fails to differentiate trichomes (Stern, 2003; Davis et al., 2007). Indeed, this region also stays trichome-free in the miR-92 loss of function line (Figures 2H, K), indicating that repression of trichomes in these cells is independent of Ubx and miR-92. The expression driven by VT42733 is also consistent with Ubx activity in T3 femurs: repression of trichomes proximally on the posterior and on the proximal dorsal-anterior region (Davis et al., 2007). Therefore the enhancer we have identified recapitulates the expression and activity of Ubx in T2 and T3 femurs (Figure 7). Interestingly, FAIRE-seq to assay the open chromatin in developing halteres and wings revealed that while the abx region is accessible there was no distinctive peak in the region of the new leg enhancer we have discovered here (McKay and Lieb, 2013; Delker et al., 2019). This suggests that while the enhancer we have identified is accessible and active in legs it is not used in the developing halteres.
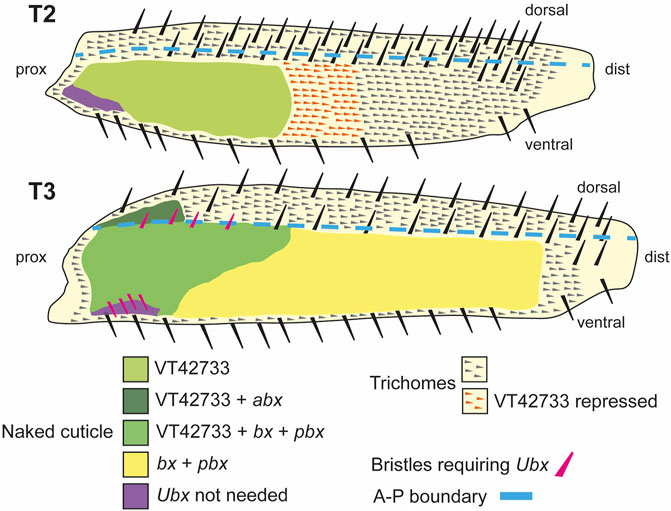
FIGURE 7. Summary of the regulation and roles of Ubx in T2 and T3 femurs. In T2 (upper scheme), Ubx expression is regulated in the proximal posterior femur by enhancer VT42733 and, together with the co-factors Exd and Hth, results in the activation of miR-92a/b and suppression of trichomes in the so called “naked valley” (light green). Repression of Ubx via VT42733 (potentially regulated by awh, C15, Dll, mirr or NK7.1) more distally in the T2 posterior femur defines the distal limit of Ubx expression and the “naked valley” (orange trichomes). In T3 (lower scheme), VT42733 and abx regulate Ubx expression in the proximal dorsal-anterior of the femur to suppress trichome development (dark green). This also requires vvl. In the posterior T3 femur, Ubx expression requires VT42733, bx and pbx proximally (medium green), and bx and pbx distally (yellow) to suppress trichomes and generate naked cuticle. Patterning of the anterior and posterior T3 femur by Ubx also requires exd and hth. The correct development of small proximal bristles on the T3 posterior femur also requires Ubx (pink bristles). In the proximal posterior-ventral femurs of both T2 and T3, trichome development is blocked independently of Ubx (purple shading). Scheme based on findings of Stern (1998), Stern (2003), Davis et al. (2007) and this study.
Davis et al. (2007) previously surveyed most of the third intron of Ubx, including the VT42733 region, for a leg enhancer (Figure 2A). However, they did not identify any regions with pupal leg activity although they found that abx is required for earlier expression during T2 development consistent with previous studies (Kerridge and Morata, 1982; Casanova et al., 1985; Peifer and Bender, 1986; Davis et al., 2007). This apparent inconsistency with our results could be explained by the different methods used to locate the enhancer. While we used reporter constructs encompassing regions of accessible chromatin in T2 pupal legs, Davis et al. (2007) studied this region using deficiencies in trans with Cbx3 and found no effect on the trichome patterning of the T2 femur. This suggests that VT42733 is able to drive expression in femur cells but removal of this region in a trans-heterozygote does not affect the trichome pattern perhaps because of compensation by additional binding sites located elsewhere in the Ubx locus. To more directly test this, it would be interesting to precisely delete the leg enhancer from the endogenous location instead of using large deficiencies of the Ubx locus that likely have pleiotropic effects and perhaps even result in pre-pupal lethality. Recent analysis of the abx enhancer resulted in similar findings to our study and those of Davis et al. (2007). Delker et al. (2019) showed that a reporter construct with a minimal region of 531 bp of the abx enhancer is able to recapitulate differential Ubx expression in proximal versus distal cells of the developing halteres (Delker et al., 2019). However, deletion of this region using CRISPR/Cas9 had no effect on this expression pattern. The authors concluded that there are likely additional binding sites elsewhere and potentially even scattered throughout the Ubx locus that contribute to its differential expression in the halteres (Delker et al., 2019).
It is clear that many of the fragments of the Ubx locus that we tested for enhancer activity, including VT42733, are active in other pupal tissues not known to normally express Ubx, for example the T1 legs. This suggests that these fragments exclude binding sites for TFs that repress Ubx in these tissues or other cis-regulatory elements like boundary elements that restrict Ubx expression to the correct locations. Ectopic expression has been observed previously with reporter constructs for regulatory regions of Scr and Ubx. abx fragments drive ectopic expression in imaginal discs that do not normally express Ubx and this has been suggested to be a consequence of their exclusion of a nearby polycomb response element (Delker et al., 2019), and potentially the variably occupied CTCF site in the third intron (Figure 2A) (Magbanua et al., 2015). Furthermore, reporter constructs for recently identified Scr enhancers that reproduce expression of this Hox gene in T1 also appear to be ectopically active in other legs where Scr is normally repressed (Eksi et al., 2018). It was suggested that these reporters contain binding sites that facilitate expression in all legs but they are missing silencer elements that normally restrict Scr to T1 (Eksi et al., 2018). This could also be the case with our Ubx leg enhancer. Alternatively, the placement of a cis-regulatory element into a different genomic location could introduce additional TF binding sites or make it accessible in tissues where it is normally inaccessible.
Candidate TFs for Ubx regulation during pupal leg development
Our identification of a T2 enhancer of Ubx allowed us to begin to decipher how this Hox gene is regulated in this developmental context and to further explore the topology of the surrounding GRN. We tested 25 TFs expressed in pupal legs with predicted binding sites in regions of accessible chromatin in VT42733, as well as the Hox co-factor Hth (Figure 4). We found that RNAi knockdown of eight of these factors affected the trichome pattern on T2 femurs (Figure 4). In contrast Giraud et al. (2021) found that only 7/117 TFs they tested had an effect on halteres, which they argued was due to robustness provided by a high dose of Ubx. Our results suggest that the trichome patterning on T2 femurs is more readily genetically perturbed, perhaps because of a lower Ubx dose distally in the femur, which might explain the extensive natural variation in this phenotype (Stern, 1998; Arif et al., 2013; Kittelmann et al., 2018).
We found that the known Hox co-factors Hth and Exd are required for Ubx function in T2 and T3 as knockdown of these TFs gave extra trichomes and bristles on dorsal-anterior and posterior femurs similar to Ubx loss of function (Stern, 1998; Stern, 2003; Davis et al., 2007) (Figure 7). hth knockdown had a stronger effect on T2 and T3 than exd RNAi, particularly on posterior trichomes and bristles. This suggests that Hth is required for Ubx mediated repression of trichomes and bristles in the proximal posterior and proximal dorsal-anterior but Exd is only required in the latter cells (Figure 7). Given the presence of putative Exd-Ubx dimer binding sites in the VT42733 sequence, this may involve Ubx autoregulation of this enhancer in proximal dorsal-anterior cells as shown for Exd-Ubx and Exd-Scr binding in other appendages (Mann, 1995; Delker et al., 2019; Feng et al., 2022). However, Exd-Ubx binding to abx represses Ubx expression proximally in halteres (Delker et al., 2019) whereas our results indicate that Exd positively regulates Ubx in T2 and T3 femurs.
Apart from Exd and Hth, the only other TF we tested that resulted in an increase in trichomes on T2 and T3 femurs when knocked down was vvl. Although vvl has no reported role in Drosophila leg disc development it is expressed in the growing appendages of other arthropods (Abzhanov and Kaufman, 2000) and our RNA-seq data show that it is expressed in Drosophila pupal legs 24 hAPF during trichome patterning (Kittelmann et al., 2018). Our results suggest that vvl represses trichomes in the T2 NV and in the dorsal-anterior of T3 perhaps by activating Ubx via the putative binding sites in the VT42733 enhancer, although it could act in parallel with or even downstream of this Hox gene.
RNAi knockdown of awh, C15, Dll, mirr and NK7.1 all resulted in an enlargement of the NV on posterior T2 femurs but had no effect on T3. These results suggest that they contribute to repressing Ubx, perhaps even directly via their predicted binding sites in the VT42733 enhancer, but again we cannot exclude the possibility that they act in parallel to this Hox gene or downstream.
It has recently been reported that Dll can act as co-factor for Scr to help regulate T1 morphology (Feng et al., 2022). Dll and Scr bind to two monomer sites separated by a short space in enhancers of Scr target genes in T1 cells (Feng et al., 2022). In T2 and T3 Dll is expressed in the coxae and distally in the femurs (Panganiban and Rubenstein, 2002; Ruiz-Losada et al., 2018). Dll could therefore also act as a Ubx co-factor to help auto-repress the expression of this Hox gene in T2 and T3 femurs. However, we did not identify any sequences like the Dll-Scr motifs in the VT42733 enhancer suggesting that if Dll does regulate Ubx in T2 and T3 it binds as a monomer to some of its eight predicted binding sites in this enhancer to repress Ubx expression. It remains possible that Dll acts downstream of Ubx to either activate trichomes distally on T2 femurs or by repressing target genes of this Hox gene that promote formation of naked cuticle. Interestingly, there is evidence that Dll represses other genes during leg development including serrate (Rauskolb, 2001).
We suggest that Dll-mediated repression of Ubx may help to promote the generation of trichomes on the distal region of the T2 femur while Ubx activates miR-92a and perhaps miR-92b more proximally to repress trichomes and generate the NV (Figure 7). However, a more detailed understanding of these regulatory interactions requires assaying whether Dll and Ubx bind directly to the Ubx and mir-92a/miR-92b enhancers, respectively.
Conclusion
Hox genes regulate fine-scale aspects of morphology as well as determining overall identity of segments along the A-P axis (Buffry and McGregor, 2022). Understanding how Hox genes are integrated into these gene regulatory networks can provide new insights into Hox gene regulation and function, and the development and evolution of morphology. We have identified a Ubx enhancer and several TFs and cofactors that may directly regulate this element as well as a downstream mechanism by which this Hox gene sculpts the fine-scale morphology of T2 and T3 femurs. Together with other studies of the regulation and role of Ubx and other Hox genes (e.g., Eksi et al., 2018; Delker et al., 2019; Paul et al., 2021; Feng et al., 2022), our findings can help to more broadly understand how Hox genes capture patterning information via their enhancers during post-embryonic development, interact in a context dependent manner with co-factors and regulate downstream targets to control cell fate and fine-scale morphology. However, a major challenge remains in identifying the likely numerous direct regulatory interactions involved to fully understand the underlying developmental gene regulatory networks.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
The project was conceived and designed by all authors. The experiments were carried out by AB and SK. Data was analysed by all authors. The manuscript was written by all authors.
Funding
AB was funded by a BBSRC DTP studentship and SK by a DFG Research Fellowship (Ki 1831/1-1).
Acknowledgments
We thank the Bloomington Drosophila Stock Center for stocks used in this study. We thank Marianne Yoth for assistance with experiments. This work relied greatly on access to information curated by FlyBase (Gramates et al., 2022). This manuscript is available as a preprint on bioRxiv (https://doi.org/10.1101/2020.06.17.152918).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1119221/full#supplementary-material
SUPPLEMENTARY FIGURE S1 (Image 1) | Ubx over-expression does not affect the anterior side of the T3 femur and requires miR-92 for the repression of ectopic trichomes. In contrast to the posterior side of the T2 femur, Ubx is unable to repress trichome development on the anterior side of T2 or T3 when over-expressed in the whole leg (A, A'', D, D''), and the femurs show no difference to controls (B, B'', C). (A′–D′) While the posterior side of a T3 femur is normally naked (A′, B′), ectopic patches of trichomes (dashed line) develop in a miR-92 loss-of-function mutant (C''). These ectopic trichomes are also not repressed by Ubx over-expression (D′).
SUPPLEMENTARY FIGURE S2 (Image 2) | (A) Expression driven by additional reporter constructs in whole pupae at 24 hAPF. Expression driven by the UbxP1 fragment is localised to the abdominal histoblasts. (B, B′) GMR31E11 drives expression in leg joints and in spots along the pupal abdomen. (C) GMR32B03 drives expression along the whole of the leg and at the periphery of the wing. (D) GMR31F12 drives expression in the abdomen in a stripe-like pattern which seems to be located in internal tissues and not the developing epithelium. (E) VT42732 does not drive expression in the pupae. (F) VT42734 drives expression in all legs. (G) e33.B drives expression in developing legs and also in the pupal antennae. (H, H′) e33.C drives some leg expression as well as in the dorsal abdomen. (I–M) The reporter constructs which did drive expression in pupal legs (31E11, 32B03, VT34, e33.B and e33.C) were functionally tested to see if they could induce trichome formation instead of naked cuticle when crossed to the trichome activating line, UAS-shaΔUTR. However, none of these lines were capable of promoting trichomes in the NV.
SUPPLEMENTARY FIGURE S3 (Image 3) | Representative T2 legs from the RNAi screen of predicted TFs. T2 proximal femurs showing the NV for each RNAi line tested.
SUPPLEMENTARY FIGURE S4 (Image 4) | TFs that significantly alter the size of the NV compared to parental control before normalisation of the measurements. When the data are normalised against the length of the T2 femur these TFs are no longer significant. (B, F, G, K, L) TFs that significantly affect the size of the NV both before and after the measurements are normalised (see Figure 4).
SUPPLEMENTARY FIGURE S5 (Image 5) | Representative T3 legs from the RNAi screen of predicted TFs. Images of T3 proximal femurs for each RNAi line tested.
SUPPLEMENTARY MATERIAL S1 (Data Sheet 1) | A list of all the fly stocks used in this study.
SUPPLEMENTARY MATERIAL S2 (Data Sheet 2) | Coordinates of reporter lines, primer sequences of reporter constructs and RedFly annotated regulatory regions for the Ubx locus.
SUPPLEMENTARY MATERIAL S3 (Data Sheet 3) | All TFBS identified by JASPAR including positions relative to open chromatin.
SUPPLEMENTARY MATERIAL S4 (Data Sheet 4) | Raw measurements of NV from all RNAi experiments including statistical significance.
References
Abzhanov, A., and Kaufman, T. C. (2000). Homologs of Drosophila appendage genes in the patterning of arthropod limbs. Dev. Biol. 227, 673–689. doi:10.1006/dbio.2000.9904
Akam, M. E., and Martinez-Arias, A. (1985). The distribution of Ultrabithorax transcripts in Drosophila embryos. EMBO J. 4, 1689–1700. doi:10.1002/j.1460-2075.1985.tb03838.x
Akam, M. (1998a). Hox genes, homeosis and the evolution of segment identity: No need for hopeless monsters. Int. J. Dev. Biol. 42, 445–451. doi:10.1387/ijdb.9654030
Akam, M. (1998b). Hox genes: From master genes to micromanagers. Curr. Biol. 8, R676–R678. doi:10.1016/s0960-9822(98)70433-6
Arif, S., Kittelmann, S., and Mcgregor, A. P. (2015). From shavenbaby to the naked valley: Trichome formation as a model for evolutionary developmental biology. Evol. Dev. 17, 120–126. doi:10.1111/ede.12113
Arif, S., Murat, S., Almudi, I., Nunes, M. D. S., Bortolamiol-Becet, D., McGregor, N. S., et al. (2013). Evolution of mir-92a underlies natural morphological variation in drosophila melanogaster. Curr. Biol. 23, 523–528. doi:10.1016/j.cub.2013.02.018
Balmert, A., Florian Bohn, H., Ditsche-Kuru, P., and Barthlott, W. (2011). Dry under water: Comparative morphology and functional aspects of air-retaining insect surfaces. J. Morphol. 272, 442–451. doi:10.1002/jmor.10921
Bender, W., Akam, M., Karch, F., Beachy, P. A., Peifer, M., Spierer, P., et al. (1983). Molecular genetics of the bithorax complex in Drosophila melanogaster. Sci. (80-. ) 221, 23–29. doi:10.1126/science.221.4605.23
Bolt, C. C., and Duboule, D. (2020). The regulatory landscapes of developmental genes. Development 147, dev171736. doi:10.1242/dev.171736
Brodu, V., Elstob, P. R., and Gould, A. P. (2002). Abdominal a specifies one cell type in Drosophila by regulating one principal target gene. Development 129, 2957–2963. doi:10.1242/dev.129.12.2957
Buffry, A. D., and McGregor, A. P. (2022). Micromanagement of Drosophila post-embryonic development by Hox genes. J. Dev. Biol. 10, 13. doi:10.3390/jdb10010013
Buffry, A. D., Mendes, C. C., and McGregor, A. P. (2016). The functionality and evolution of eukaryotic transcriptional enhancers. Adv. Genet. 96, 143–206. doi:10.1016/bs.adgen.2016.08.004
Casanova, J., Sánchez-Herrero, E., and Morata, G. (1985). Prothoracic transformation and functional structure of the Ultrabithorax gene of Drosophila. Cell 42, 663–669. doi:10.1016/0092-8674(85)90123-0
Castelli-Gair, J., and Akam, M. (1995). How the Hox gene Ultrabithorax specifies two different segments: The significance of spatial and temporal regulation within metameres. Development 121, 2973–2982. doi:10.1242/dev.121.9.2973
Chanut-Delalande, H., Fernandes, I., Roch, F., Payre, F., and Plaza, S. (2006). Shavenbaby couples patterning to epidermal cell shape control. PLoS Biol. 4, e290. doi:10.1371/journal.pbio.0040290
Crocker, J., Abe, N., Rinaldi, L., McGregor, A. P., Frankel, N., Wang, S., et al. (2015). Low affinity binding site clusters confer HOX specificity and regulatory robustness. Cell 160, 191–203. doi:10.1016/j.cell.2014.11.041
Davis, G. K., Srinivasan, D. G., Wittkopp, P. J., and Stern, D. L. (2007). The function and regulation of Ultrabithorax in the legs of Drosophila melanogaster. Dev. Biol. 308, 621–631. doi:10.1016/j.ydbio.2007.06.002
Delker, R. K., Ranade, V., Loker, R., Voutev, R., and Mann, R. S. (2019). Low affinity binding sites in an activating CRM mediate negative autoregulation of the Drosophila Hox gene Ultrabithorax. PLoS Genet. 15, e1008444. doi:10.1371/journal.pgen.1008444
Delon, I., Chanut-Delalande, H., and Payre, F. (2003). The Ovo/Shavenbaby transcription factor specifies actin remodelling during epidermal differentiation in Drosophila. Mech. Dev. 120, 747–758. doi:10.1016/S0925-4773(03)00081-9
Diaz-de-la-Loza, M.-D.-C., Loker, R., Mann, R. S., and Thompson, B. J. (2020). Control of tissue morphogenesis by the HOX gene Ultrabithorax. Development 147, dev184564. doi:10.1242/dev.184564
Ditsche-Kuru, P., Schneider, E. S., Melskotte, J. E., Brede, M., Leder, A., and Barthlott, W. (2011). Superhydrophobic surfaces of the water bug notonecta glauca: A model for friction reduction and air retention. Beilstein J. Nanotechnol. 2, 137–144. doi:10.3762/bjnano.2.17
Eksi, S. E., Barmina, O., McCallough, C. L., Kopp, A., and Orenic, T. V. (2018). A Distalless-responsive enhancer of the Hox gene Sex combs reduced is required for segment- and sex-specific sensory organ development in Drosophila. PLoS Genet. 14, e1007320. doi:10.1371/journal.pgen.1007320
Estacio-Gómez, A., Moris-Sanz, M., Schäfer, A. K., Perea, D., Herrero, P., and Díaz-Benjumea, F. J. (2013). Bithorax-complex genes sculpt the pattern of leucokinergic neurons in the Drosophila central nervous system. Dev 140, 2139–2148. doi:10.1242/dev.090423
Feng, S., Rastogi, C., Loker, R., Glassford, W. J., Tomas Rube, H., Bussemaker, H. J., et al. (2022). Transcription factor paralogs orchestrate alternative gene regulatory networks by context-dependent cooperation with multiple cofactors. Nat. Commun. 13, 3808. doi:10.1038/s41467-022-31501-2
Fornes, O., Castro-Mondragon, J. A., Khan, A., van der Lee, R., Zhang, X., Richmond, P. A., et al. (2020). JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92. doi:10.1093/nar/gkz1001
Frankel, N. S., Erezyilmaz, D. F., McGregor, A. P., Wang, S., Payre, F., and Stern, D. L. (2011). Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature 474, 598–603. doi:10.1038/nature10200
Frankel, N., Wang, S., and Stern, D. L. (2012). Conserved regulatory architecture underlies parallel genetic changes and convergent phenotypic evolution. Proc. Natl. Acad. Sci. U. S. A. 109, 20975–20979. doi:10.1073/pnas.1207715109
Giraud, G., Paul, R., Duffraisse, M., Khan, S., Shashidhara, L. S., and Merabet, S. (2021). Developmental robustness: The haltere case in Drosophila. Front. Cell Dev. Biol. 9, 713282. doi:10.3389/fcell.2021.713282
Gramates, L. S., Agapite, J., Attrill, H., Calvi, B. R., Crosby, M. A., Dos Santos, G., et al. (2022). Fly base: A guided tour of highlighted features. Genetics 220, iyac035. doi:10.1093/genetics/iyac035
Halfon, M. S. (2019). Studying transcriptional enhancers: The founder fallacy, validation creep, and other biases. Trends Genet. 35, 93–103. doi:10.1016/j.tig.2018.11.004
Hombría, J. C.-G., and Lovegrove, B. (2003). Beyond homeosis--HOX function in morphogenesis and organogenesis. Differentiation 71, 461–476. doi:10.1046/j.1432-0436.2003.7108004.x
Irvine, K. D., Botas, J., Jha, S., Mann, R. S., and Hogness, D. S. (1993). Negative autoregulation by Ultrabithorax controls the level and pattern of its expression. Development 117, 387–399. doi:10.1242/dev.117.1.387
Irvine, K. D., Helfand, S. L., and Hogness, D. S. (1991). The large upstream control region of the Drosophila homeotic gene Ultrabithorax. Development 111, 407–424. doi:10.1242/dev.111.2.407
Jenett, A., Rubin, G. M., Ngo, T.-T. B., Shepherd, D., Murphy, C., Dionne, H., et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001. doi:10.1016/j.celrep.2012.09.011
Jindal, G. A., and Farley, E. K. (2021). Enhancer grammar in development, evolution, and disease: Dependencies and interplay. Dev. Cell 56, 575–587. doi:10.1016/j.devcel.2021.02.016
Kannan, R., Berger, C., Myneni, S., Technau, G. M., and Shashidhara, L. S. (2010). Abdominal-A mediated repression of Cyclin E expression during cell-fate specification in the Drosophila central nervous system. Mech. Dev. 127, 137–145. doi:10.1016/j.mod.2009.09.008
Kerridge, S., and Morata, G. (1982). Developmental effects of some newly induced Ultrabithorax alleles of Drosophila. J. Embryol. Exp. Morphol. 68, 211–234. doi:10.1242/dev.68.1.211
Kittelmann, S., Buffry, A. D., Franke, F. A., Almudi, I., Yoth, M., Sabaris, G., et al. (2018). Gene regulatory network architecture in different developmental contexts influences the genetic basis of morphological evolution. PLoS Genet. 14, e1007375. doi:10.1371/journal.pgen.1007375
Kittelmann, S., Preger-Ben Noon, E., McGregor, A. P., and Frankel, N. (2021). A complex gene regulatory architecture underlies the development and evolution of cuticle morphology in Drosophila. Curr. Opin. Genet. Dev. 69, 21–27. doi:10.1016/j.gde.2021.01.003
Krumlauf, R. (2018). Hox genes, clusters and collinearity. Int. J. Dev. Biol. 62, 659–663. doi:10.1387/ijdb.180330rr
Lewis, E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565–570. doi:10.1038/276565a0
Little, J. W., Byrd, C. A., and Brower, D. L. (1990). Effect of abx, bx and pbx mutations on expression of homeotic genes in Drosophila larvae. Genetics 124, 899–908. doi:10.1093/genetics/124.4.899
Loker, R., Sanner, J. E., and Mann, R. S. (2021). Cell-type-specific Hox regulatory strategies orchestrate tissue identity. Curr. Biol. 31, 4246–4255.e4. doi:10.1016/j.cub.2021.07.030
Maeda, R. K., and Karch, F. (2006). The ABC of the BX-C: The bithorax complex explained. Development 133, 1413–1422. doi:10.1242/dev.02323
Magbanua, J. P., Runneburger, E., Russell, S., and White, R. (2015). A variably occupied CTCF binding site in the Ultrabithorax gene in the Drosophila bithorax complex. Mol. Cell. Biol. 35, 318–330. doi:10.1128/mcb.01061-14
Mann, R. S. (1995). The specificity of homeotic gene function. BioEssays 17, 855–863. doi:10.1002/bies.950171007
McGinnis, W., and Krumlauf, R. (1992). Homeobox genes and axial patterning. Cell 68, 283–302. doi:10.1016/0092-8674(92)90471-n
McGregor, A. P., Orgogozo, V., Delon, I., Zanet, J., Srinivasan, D. G., Payre, F., et al. (2007). Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448, 587–590. doi:10.1038/nature05988
McKay, D. J., and Lieb, J. D. (2013). A common set of DNA regulatory elements shapes Drosophila appendages. Dev. Cell 27, 306–318. doi:10.1016/j.devcel.2013.10.009
Menoret, D., Santolini, M., Fernandes, I., Spokony, R., Zanet, J., Gonzalez, I., et al. (2013). Genome-wide analyses of Shavenbaby target genes reveals distinct features of enhancer organization. Genome Biol. 14, R86. doi:10.1186/gb-2013-14-8-r86
Morata, G., and Kerridge, S. (1981). Sequential functions of the bithorax complex of Drosophila. Nature 290, 778–781. doi:10.1038/290778a0
Müller, J., and Bienz, M. (1991). Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 10, 3147–3155. doi:10.1002/j.1460-2075.1991.tb04876.x
Panganiban, G., and Rubenstein, J. L. R. (2002). Developmental functions of the Distal-less/Dlx homeobox genes. Development 129, 4371–4386. doi:10.1242/dev.129.19.4371
Paul, R., Giraud, G., Domsch, K., Duffraisse, M., Marmigère, F., Khan, S., et al. (2021). Hox dosage contributes to flight appendage morphology in Drosophila. Nat. Commun. 12, 2892. doi:10.1038/s41467-021-23293-8
Pavlopoulos, A., and Akam, M. (2011). Hox gene Ultrabithorax regulates distinct sets of target genes at successive stages of Drosophila haltere morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 108, 2855–2860.
Pearson, J. C., Lemons, D., and McGinnis, W. (2005). Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6, 893–904. doi:10.1038/nrg1726
Peifer, M., and Bender, W. (1986). The anterobithorax and bithorax mutations of the bithorax complex. EMBO J. 5, 2293–2303. doi:10.1002/j.1460-2075.1986.tb04497.x
Pfeiffer, B. D., Jenett, A., Hammonds, A. S., Ngo, T.-T. B., Misra, S., Murphy, C., et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 105, 9715–9720. doi:10.1073/pnas.0803697105
Pirrotta, V., Chan, C. S., McCabe, D., and Qian, S. (1995). Distinct parasegmental and imaginal enhancers and the establishment of the expression pattern of the Ubx gene. Genetics 141, 1439–1450. doi:10.1093/genetics/141.4.1439
Preger-Ben Noon, E., Davis, F. P., and Stern, D. L. (2016). Evolved repression overcomes enhancer robustness. Dev. Cell 39, 572–584. doi:10.1016/j.devcel.2016.10.010
Rauskolb, C. (2001). The establishment of segmentation in the Drosophila leg. Development 128, 4511–4521. doi:10.1242/dev.128.22.4511
Rivera, J., Keränen, S. V. E., Gallo, S. M., and Halfon, M. S. (2019). REDfly: The transcriptional regulatory element database for Drosophila. Nucleic Acids Res. 47, D828–D834. doi:10.1093/nar/gky957
Roch, F., and Akam, M. (2000). Ultrabithorax and the control of cell morphology in Drosophila halteres. Development 127, 97–107. doi:10.1242/dev.127.1.97
Ruiz-Losada, M., Blom-Dahl, D., Córdoba, S., and Estella, C. (2018). Specification and patterning of Drosophila appendages. J. Dev. Biol. 6, 17. doi:10.3390/jdb6030017
Schertel, C., Rutishauser, T., Förstemann, K., and Basler, K. (2012). Functional characterization of Drosophila microRNAs by a novel in vivo library. Genetics 192, 1543–1552. doi:10.1534/genetics.112.145383
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi:10.1038/nmeth.2019
Schubiger, G., Schubiger, M., and Sustar, A. (2012). The three leg imaginal discs of Drosophila: “Vive la différence”. Dev. Biol. 369, 76–90. doi:10.1016/j.ydbio.2012.05.025
Shroff, S., Joshi, M., and Orenic, T. V. (2007). Differential Delta expression underlies the diversity of sensory organ patterns among the legs of the Drosophila adult. Mech. Dev. 124, 43–58. doi:10.1016/j.mod.2006.09.004
Simon, J., Peifer, M., Bender, W., and O’Connor, M. (1990). Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 9, 3945–3956. doi:10.1002/j.1460-2075.1990.tb07615.x
Stern, D. L. (1998). A role of Ultrabithorax in morphological differences between Drosophila species. Nature 396, 463–466. doi:10.1038/24863
Stern, D. L., and Frankel, N. (2013). The structure and evolution of cisregulatory regions: The shavenbaby story. Philos. Trans. R. Soc. B Biol. Sci. 368, 20130028. doi:10.1098/rstb.2013.0028
Stern, D. L. (2003). The Hox gene Ultrabithorax modulates the shape and size of the third leg of Drosophila by influencing diverse mechanisms. Dev. Biol. 256, 355–366. doi:10.1016/S0012-1606(03)00035-6
Struhl, G. (1982). Genes controlling segmental specification in the Drosophila thorax. Proc. Natl. Acad. Sci. U. S. A. 79, 7380–7384. doi:10.1073/pnas.79.23.7380
Sucena, E., Delon, I., Jones, I., Payre, F., and Stern, D. L. (2003). Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424, 935–938. doi:10.1038/nature01768
Sucena, É., and Stern, D. L. (2000). Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc. Natl. Acad. Sci. U. S. A. 97, 4530–4534. doi:10.1073/pnas.97.9.4530
Weatherbee, S. D., Halder, G., Kim, J., Hudson, A., and Carroll, S. (1998). Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev. 12, 1474–1482. doi:10.1101/gad.12.10.1474
White, R. A. H., and Akam, M. E. (1985). Contrabithorax mutations cause inappropriate expression of Ultrabithorax products in Drosophila. Nature 318, 567–569. doi:10.1038/318567a0
White, R. A. H., and Wilcox, M. (1985). Distribution of Ultrabithorax proteins in Drosophila. EMBO J. 4, 2035–2043. doi:10.1002/j.1460-2075.1985.tb03889.x
White, R. A. H., and Wilcox, M. (1984). Protein products of the bithorax complex in Drosophila. Cell 163, 163–171. doi:10.1016/0092-8674(84)90202-2
Keywords: Drosophila, Hox genes, gene regulation, evolution, enhancers, development, Ultrabithorax
Citation: Buffry AD, Kittelmann S and McGregor AP (2023) Characterisation of the role and regulation of Ultrabithorax in sculpting fine-scale leg morphology. Front. Cell Dev. Biol. 11:1119221. doi: 10.3389/fcell.2023.1119221
Received: 08 December 2022; Accepted: 20 January 2023;
Published: 13 February 2023.
Edited by:
Markus Friedrich, Wayne State University, United StatesReviewed by:
Ralf Janssen, Uppsala University, SwedenIgnacio Maeso, Faculty of Biology, University of Barcelona, Spain
Thomas Williams, University of Dayton, United States
Copyright © 2023 Buffry, Kittelmann and McGregor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alistair P. McGregor, alistair.mcgregor@durham.ac.uk
 Alexandra D. Buffry
Alexandra D. Buffry Sebastian Kittelmann
Sebastian Kittelmann Alistair P. McGregor
Alistair P. McGregor