Abstract
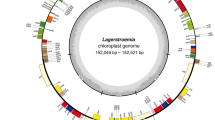
The subgenus Anguinum (G. Don. ex W.D.J. Koch) N. Friesen disjunctly distributes in the high mountains from south-western Europe to eastern Asia and in northeastern North America. Due to interspecies hybridization, introgression and incomplete lineage sorting, the detection of interspecies differences within Anguinum is notoriously challenging. Here we report the complete chloroplast (cp) genome sequences of A. nanodes Airy-Shaw, A. ovalifolium Hand.-Mazz., A. ovalifolium var. leuconeurum J.M. Xu and A. victorialis L. from Anguinum and A. cyathophorum Bur. & Franch from subgenus Cyathophora (R.M. Fritsch) R.M. Fritsch as outgroup, four of which were first reported. We compared them with cp genomes of A. cepa L., A. obliquum L., A. prattii C.H. Wright. The Anguinum cp genomes ranged from 153,674 to 155,055 bp in length. Each cp genome contained 131 unigenes, consisting of 85 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The eight Allium L. cp genomes exhibited significant differences at the SC/IR junction regions. Fourteen cpDNA markers with most variable sites were identified as mutational hotspots, and simple sequence repeats (SSRs) and long repeats were also identified. Three single-copy genes (accD, rps14, rpl33) may be under a great selection pressure indicated by positive selection analysis. The plastome-based phylogeny indicated a monophyletic position of the subgenus Anguinum, which was consistent with previous phylogenetic studies. Overall, the availability of these complete cp genomes provides valuable information for further studies of population genetics and investigation of the evolution in the genus Allium.






Similar content being viewed by others
REFERENCES
Seregin, A.P., Anačkov, G., and Friesen, N., Molecular and morphological revision of the Allium saxatile group (Amaryllidaceae): geographical isolation as the driving force of underestimated speciation, Bot. J. Linn. Soc., 2015, vol. 178, pp. 67—101.
Li, Q.Q., Zhou, S.D., Huang, D.Q., et al., Molecular phylogeny, divergence time estimates and historical biogeography within one of the world’s largest monocot genera, AoB Plants, 2016, vol. 8. plw041.
Fay, M.F. and Chase, M.W., Resurrection of Themidaceae for the Brodiaea alliance, and recircumscription of Alliaceae, Amaryllidaceae and Agapanthoideae, Taxon, 1996, vol. 45, pp. 441—451.
APG III, An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III, Bot. J. Linn. Soc., 2009, vol. 161, pp. 105—121.
Chase, M.V., Reveal, J.L., and Fay, M.F., A subfamilial classification for the expanded asparagalean families Amaryllidaceae, Asparagaceae and Xanthorrhoeaceae, Bot. J. Linn. Soc., 2009, vol.161, pp. 132—136.
Fritsch, R.M., Taxonomy of the genus Allium: contribution from IPK Gatersleben, Herbertia, 2001, vol. 56, pp. 19—50.
Fritsch, R.M. and Friesen, N., Evolution, Domestication and Taxonomy, Allium Crop Science: Recent Advances, New York: CAB International, 2002, pp. 5—30.
Friesen, N., Fritsch, R.M., and Blattner, F.R., Phylogeny and new intrageneric classification of Allium (Alliaceae) based on nuclear ribosomal DNA ITS sequences, Aliso: J. Syst. Evol. Bot., 2006, vol. 22, pp. 372—395.
Li, Q.Q., Zhou, S.D., He, X.J., et al., Phylogeny and biogeography of Allium (Amaryllidaceae: Allieae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps16 sequences, focusing on the inclusion of species endemic to China, Ann. Bot., 2010, vol. 106, pp. 709—733.
Choi, H.J., Giussani, L.M., Jang, C.G., et al., Systematics of disjunct northeastern Asian and northern North American Allium (Amaryllidaceae), Botany, 2012, vol. 90, pp. 491—508.
Dahal, S., Sharma, T.P., and Borthakur, S.K., Database on medicinal plants of Tamze Medicinal Plants Conservation Area (MPCA) of Sikkim Himalaya, India, NeBIO—Int. J. Environ. Biodiversity, 2017, vol. 8, pp. 45—56.
Pastor, J. and Valdes, B., Bulb structure in some species of Allium (Liliaceae) of Iberian Peninsula, Ann. Mus. Goulandris., 1985, vol. 7, pp. 249—261.
Kruse, J., Rasterelektronenmikroskopische Untersuchungen an Samen der Gattung Allium L.: 4, Feddes Repertorium, 1994, vol. 105, pp. 457—471.
Fritsch, R.M., Zur Wurzelanatomie in der Gattung Allium L. (Alliaceae), Beitr. Biol. Pflanz., 1992, vol. 67, pp. 129—160.
Druselmann, S., Vergleichende Untersuchungen an Vertretern der Alliaceae Agardh: 1. Morphologie der Keimpflanzen der Gattung Allium L., Flora, 1992, vol. 186, pp. 37—52.
Jing, W.C., Xu, J.M., and Yang, L., A study on cytotaxonomy of sect. Anguinum of Allium, Acta. Phytotaxon. Sin., 1999, vol. 37, pp. 20—34.
Herden, T., Hanelt, P., and Friesen, N., Phylogeny of Allium L. subgenus Anguinum (G. Don. ex W.D.J. Koch) N. Friesen (Amaryllidaceae), Mol. Phylogenet. Evol., 2016, vol. 95, pp. 79—93.
Straub, S.C.K., Parks, M., Weitemier, K., et al., Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics, Am. J. Bot., 2012, vol. 99, pp. 349—364.
Zeng, L.P., Zhang, Q., Sun, R.R., et al., Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times, Nat. Commun., 2014, vol. 5, p. 12.
Barrett, C.F., Baker, W.J., Comer, J.R., et al., Plastid genomes reveal support for deep phylogenetic relationships and extensive rate variation among palms and other commelinid monocots, New. Phytol., 2016, vol. 209, pp. 855—870.
Twyford, A.D. and Ness, R.W., Strategies for complete plastid genome sequencing, Mol. Ecol. Resour., 2017, vol. 17, pp. 858—868.
Zhou, T., Chen, C., Wei, Y., et al., Comparative transcriptome and chloroplast genome analyses of two related Dipteronia species, Front. Plant. Sci., 2016, vol. 7, p. 1512.
Zhu, A.D., Guo, W.H., Gupta, S., et al., Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates, New. Phytol., 2016, vol. 209, pp. 1747—1756.
Xu, W.Q., Losh, J., Chen, C., et al., Comparative genomics of figworts (Scrophularia, Scrophulariaceae), with implications for the evolution of Scrophularia and Lamiales, J. Syst. Evol., 2018, vol. 999, pp. 1—11.
Fu, C.N., Li, H.T., Milne, R., et al., Comparative analyses of plastid genomes from fourteen Cornales species: inferences for phylogenetic relationships and genome evolution, BMC Genomics, 2017, vol. 18, p. 956.
Ni, Z.X., Ye, Y.J., Bai, T.D., et al., Complete chloroplast genome of Pinus massoniana (Pinaceae): gene rearrangements, loss of ndh genes, and short inverted repeats contraction, expansion, Molecules, 2017, vol. 22, p. 1528.
Huang, D.I., Hefer, C.A., Kolosova, N., et al., Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae), New. Phytol., 2014, vol. 204, pp. 693—703.
Yurina, N.P. and Odintsova, M.S., Comparative structural organization of plant chloroplast and mitochondrial genomes, Russ. J. Genet., 1998, vol. 34, pp. 1—16.
Jansen, R.K., Raubeson, L.A., Boore, J.L., et al., Methods for obtaining and analyzing whole chloroplast genome sequences, Methods Enzymol., 2005, vol. 395, pp. 348—384.
Goulding, S.E., Olmstead, R.G., Morden, C.W., and Wolfe, K.H., Ebb and flow of the chloroplast inverted repeat, Mol. Gen. Genet., 1996, vol. 252, pp. 195—206.
Wang, R.J., Cheng, C.L., Chang, C.C., et al., Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots, BMC Evol. Biol., 2008, vol. 8, p. 36.
Wicke, S., Schaferhoff, B., Depamphilis, C.W., and Müller, K.F., Disproportional plastome-wide increase of substitution rates and relaxed purifying selection in genes of Carnivorous Lentibulariaceae, Mol. Biol. Evol., 2014, vol. 31, pp. 529—545.
Downie, S.R. and Jansen, R.K., A comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions, Syst. Bot., 2015, vol. 40, pp. 336—351.
Wu, C.S. and Chaw, S.M., Evolutionary stasis in cycad plastomes and the first case of plastome GC-biased gene conversion, Genome Biol. Evol., 2015, vol. 7, pp. 2000—2009.
Keeling, P.J. and Palmer, J.D., Horizontal gene transfer in eukaryotic evolution, Nat. Rev. Genet., 2008, vol. 9, p. 605.
Daniell, H., Wurdack, K.J., Kanagaraj, A., et al., The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and the evolution of atpF in Malpighiales: RNA editing and multiple losses of a group II intron, Theor. Appl. Genet., 2008, vol. 116, p. 723.
Peredo, E.L., King, U.M., and Les, D.H., The plastid genome of Najas flexilis: adaptation to submersed environments is accompanied by the complete loss of the NDH complex in an aquatic angiosperm, PLoS One, 2013, vol. 8. e68591
Doyle, J.J., A rapid DNA isolation procedure for small quantities of fresh leaf tissue, Phytochem. Bull., 1987, vol. 19, pp. 11—15.
Zerbino, D.R. and Birney, E., Velvet: algorithms for de novo short read assembly using de Bruijn graphs, Genome Res., 2008, vol. 18, pp. 821—829.
Schulz, M.H., Zerbino, D.R., Vingron, M., and Birney, E., Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels, Bioinformatics, 2012, vol. 28, pp. 1086—1092.
Kearse, M., Moir, R., Wilson, A., et al., Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data, Bioinformatics, 2012, vol. 28, pp. 1647—1649.
Huang, D.I. and Cronk, Q.C.B., Plann: a command-line application for annotating plastome sequences, Appl. Plant. Sci., 2015, vol. 3, p. 1500026.
Lohse, M., Drechsel, O. and Bock, R., OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes, Curr. Genet., 2007, vol. 52, pp. 267—274.
Kurtz, S., Choudhuri, J.V., Ohlebusch, E., et al., REPuter: the manifold applications of repeat analysis on a genomic scale, Nucleic Acids Res., 2001, vol. 29, pp. 4633—4642.
Li, Q. and Wan, J.M., SSRHunter: development of a local searching software for SSR sites, Yi Chuan, 2005, vol. 27, pp. 808—810.
Katoh, K., Misawa, K., Kei-ichi, K., and Miyata, T., MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform, Nucleic Acids Res., 2002, vol. 30, pp. 3059—3066.
Castresana, J., Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis, Mol. Biol. Evol., 2000, vol. 17, pp. 540—552.
Stamatakis, A., RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies, Bioinformatics, 2014, vol. 30, pp. 1312—1313.
Li, L., Stoeckert, C.J., and Roos, D.S., OrthoMCL: identification of ortholog groups for eukaryotic genomes, Genome Res., 2003, vol. 13, pp. 2178—2189.
Löytynoja, A., Phylogeny-aware alignment with PRANK, in Multiple Sequence Alignment Methods, Totowa: Humana, 2014, pp. 155—170.
Yang, Z.H., PAML 4: phylogenetic analysis by maximum likelihood, Mol. Biol. Evol., 2007, vol. 24, pp. 1586—1591.
Huotari, T. and Korpelainen, H., Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes, Gene, 2012, vol. 508, pp. 96—105.
Plunkett, G.M. and Downie, S.R., Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae, Syst. Bot., 2000, vol. 25, pp. 648—667.
Goremykin, V.V., Nikiforova, S.V., Biggs, P.J., et al., The evolutionary root of flowering plants, Syst. Biol., 2012, vol. 62, pp. 50—61.
Ma, J., Yang, B.X., Zhu, W., et al., The complete chloroplast genome sequence of Mahonia bealei (Berberidaceae) reveals a significant expansion of the inverted repeat and phylogenetic relationship with other angiosperms, Gene, 2013, vol. 528, pp. 120—131.
Weng, M.L., Blazier, J.C., Govindu, M., and Jansen, R.K., Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates, Mol. Biol. Evol., 2014, vol. 31, pp. 645—659.
Dugas, D.V., Hernandez, D., Koenen, E.J.M., et al., Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions, and accelerated rate of evolution in clpP, Sci. Rep., 2015, vol. 5, p. 16958.
Song, Y., Dong, W.P., Liu, B., et al., Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae, Front. Plant Sci., 2015, vol. 6, p. 662.
Wolf, P.G., Roper, J.M. and Duffy, A.M., The evolution of chloroplast genome structure in ferns, Genome, 2010, vol. 53, pp. 731—738.
Jansen, R.K., Cai, Z., Raubeson, L.A., et al., Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns, Proc. Natl. Acad. Sci., 2007, vol. 104, pp. 19369—19374.
Green, B.R., Chloroplast genomes of photosynthetic eukaryotes, Plant J., 2011, vol. 66, pp. 34—44.
Blazier, J.C., Guisinger, M.M., and Jansen, R.K., Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae), Plant Mol. Biol., 2011, vol. 76, pp. 263—272.
Ni, L.H., Zhao, Z.L., Xu, H.X., et al., Chloroplast genome structures in Gentiana (Gentianaceae), based on three medicinal alpine plants used in Tibetan herbal medicine, Curr. Genet., 2017, vol. 63, pp. 241—252.
Kawaguchi, M., Hiroi, J., Miya, M., et al., Intron-loss evolution of hatching enzyme genes in Teleostei, BMC Evol. Biol., 2010, vol. 10, p. 260.
Ivanova, Z., Sablok, G., Daskalova, E., et al., Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response, Front. Plant Sci., 2017, vol. 8, p. 204.
Gemayel, R., Cho, J., Boeynaems, S., and Verstrepen, K.J., Beyond junk-variable tandem repeats as facilitators of rapid evolution of regulatory and coding sequences, Genes, 2012, vol. 3, pp. 461—480.
Voronova, A., Belevich, V., Jansons, A., and Rungis, D., Stress-induced transcriptional activation of retrotransposon-like sequences in the Scots pine (Pinus sylvestris L.) genome, Tree Genet. Genomes, 2014, vol. 10, pp. 937—951.
Wu, Z., Tembrock, L.R., and Ge, S., Are differences in genomic data sets due to true biological variants or errors in genome assembly: an example from two chloroplast genomes, PLoS One, 2015, vol. 10. e0118019
Zhang, Y.J., Du, L.W., Liu, A., et al., The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses, Front. Plant Sci., 2016, vol. 7, p. 306.
Daniell, H., Lin, C.S., Yu M., and Chang, W.J., Chloroplast genomes: diversity, evolution, and applications in genetic engineering, Genome Biol., 2016, vol. 17, p. 134.
Williams, A.V., Miller, J.T., Small, I., et al., Integration of complete chloroplast genome sequences with small amplicon datasets improves phylogenetic resolution in Acacia, Mol. Phylogenet. Evol., 2016, vol. 96, pp. 1—8.
Tonti-Filippini, J., Nevill, P.G., Dixon, K., and Small, I., What can we do with 1000 plastid genomes?, Plant J., 2017, vol. 90, pp. 808—818.
Stebbins, G.L., Chromosomal Evolution in Higher Plants, London: Science Reviews, 1971, pp. 270—272.
Shaw, J., Lickey, E.B., Schilling, E.E., and Small, R.L., Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III, Am. J. Bot., 2007, vol. 94, pp. 275—288.
Dong, W.P., Liu, J., Yu, J., et al., Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding, PLoS One, 2012, vol. 7. e35071
Kim, K.J. and Jansen, R.K., ndhF sequence evolution and the major clades in the sunflower family, Proc. Natl. Acad. Sci., 1995, vol. 92, pp. 10379—10383.
Müller, K. and Borsch, T., Systematics of Utricularia (Lentibulariaceae) and molecular evolution of the trnK intron in a lineage with high mutational rates, Plant Syst. Evol., 2005, vol. 250, pp. 39—67.
Müller, K. and Borsch, T., Phylogenetics of Amaranthaceae using matK/trnK sequence data-evidence from parsimony, likelihood and Bayesian approaches, Ann. Mo. Bot. Gard., 2005, vol. 92, pp. 66—102.
Taberlet, P., Coissac, E., Pompanon, F., et al., Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding, Nucleic Acids Res., 2006, vol. 35. e14
Wanke, S., Jaramillo, M.A., Borsch, T., et al., Evolution of Piperales—matK gene and trnK intron sequence data reveal lineage specific resolution contrast, Mol. Phylogenet. Evol., 2007, vol. 42, pp. 477—497.
Group, C.P.W., Hollingsworth, P.M., Forrest, L.L., et al., A DNA barcode for land plants, Proc. Natl. Acad. Sci. 2009, vol. 106, pp. 12794—12797.
Dong, W.P., Xu, C., Li, C.H., et al., ycf1, the most promising plastid DNA barcode of land plants, Sci. Rep., 2015, vol. 5, p. 8348.
Sasaki, Y. and Nagano, Y., Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding, Biosci. Biotechnol. Biochem., 2004, vol. 68, pp. 1175—1184.
Figueroa, P., Gómez, I., Holuigue, L., et al., Transfer of rps14 from the mitochondrion to the nucleus in maize implied integration within a gene encoding the iron—sulphur subunit of succinate dehydrogenase and expression by alternative splicing, Plant J., 1999, vol. 18, pp. 601—609.
Rogalski, M., Schöttler, M.A., Thiele, W., et al., Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions, Plant Cell, 2008, vol. 20, pp. 2221—2237.
ACKNOWLEDGMENTS
The author was grateful to the opened cp genome data from public databases. The author thanked Chuan Xie, Jin-Bo Tan, Fu-Min Xie and Jun-Pei Chen for the help of sequence analysis. We declare that there are no known conflicts of interest associated with this publication. We undertake that the work is our original work and the manuscript have not been published elsewhere or simultaneously submitted to other journals.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 31872647, 31570198), the Specimen Platform of China, Teaching Specimen’s sub-platform (Available website: http://mnh.scu.edu.cn/), the Science and Technology Basic Work (grant no. 2013FY112100).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Rights and permissions
About this article
Cite this article
Jin, F.Y., Y, X., Xie, D.F. et al. Comparative Complete Chloroplast Genome Analyses and Contribution to the Understanding of Chloroplast Phylogeny and Adaptive Evolution in Subgenus Anguinum. Russ J Genet 55, 872–884 (2019). https://doi.org/10.1134/S1022795419070081
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795419070081