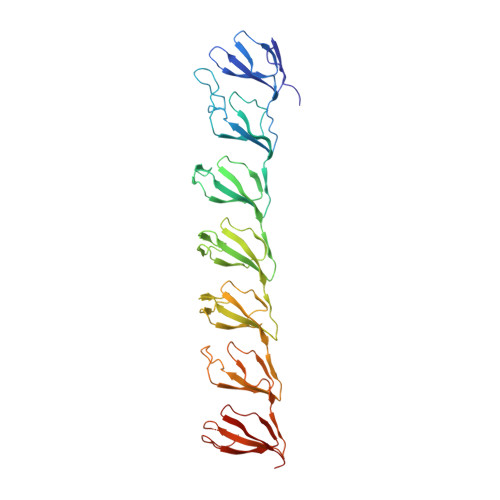The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP
Querol-Audi, J., Casanas, A., Uson, I., Luque, D., Caston, J.R., Fita, I., Verdaguer, N.(2009) EMBO J 28: 3450-3457
- PubMed: 19779459
- DOI: https://doi.org/10.1038/emboj.2009.274
- Primary Citation of Related Structures:
3GF5, 3GNF, 3GNG - PubMed Abstract:
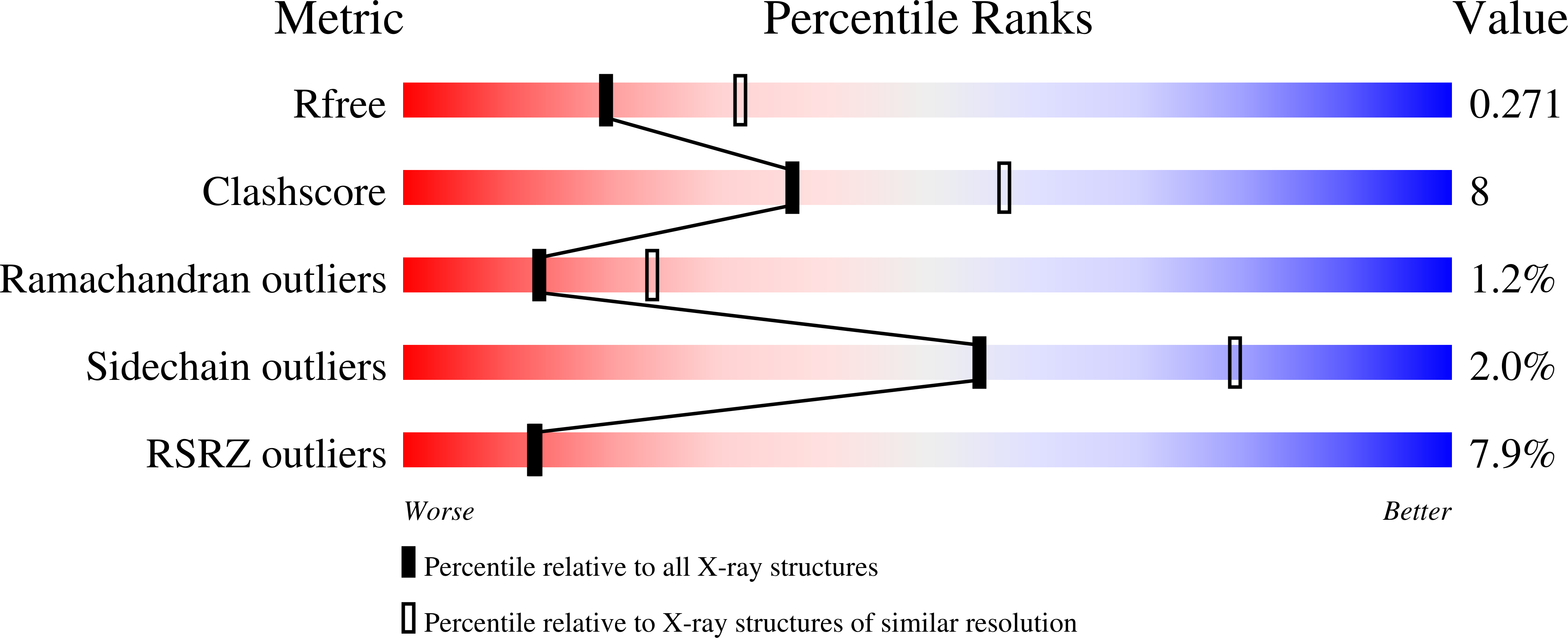
Vaults are ubiquitous ribonucleoprotein complexes involved in a diversity of cellular processes, including multidrug resistance, transport mechanisms and signal transmission. The vault particle shows a barrel-shaped structure organized in two identical moieties, each consisting of 39 copies of the major vault protein MVP. Earlier data indicated that vault halves can dissociate at acidic pH. The crystal structure of the vault particle solved at 8 A resolution, together with the 2.1-A structure of the seven N-terminal domains (R1-R7) of MVP, reveal the interactions governing vault association and provide an explanation for a reversible dissociation induced by low pH. The structural comparison with the recently published 3.5 A model shows major discrepancies, both in the main chain tracing and in the side chain assignment of the two terminal domains R1 and R2.
Organizational Affiliation:
Department of Structural Biology, Institut de Biología Molecular de Barcelona (CSIC), Baldiri i Reixac, Barcelona, Spain.