Abstract
SMIFH2 is a small molecule inhibitor of the formin family of cytoskeletal regulators that was originally identified in a screen for suppression of actin polymerization induced by the mouse formin Diaphanous 1 (mDia1). Despite widespread use of this compound, it is unknown whether SMIFH2 inhibits all human formins. Additionally, the nature of protein/inhibitor interactions remains elusive. We assayed SMIFH2 against human formins representing six of the seven mammalian classes and found inhibitory activity against all formins tested. We synthesized a panel of SMIFH2 derivatives and found that, while many alterations disrupt SMIFH2 activity, substitution of an electron-donating methoxy group in place of the bromine along with halogenation of the furan ring increases potency by approximately five-fold. Similar to SMIFH2, the active derivatives are also pan-inhibitors for the formins tested. This result suggests that while potency can be improved, the goal of distinguishing between highly conserved FH2 domains may not be achievable using the SMIFH2 scaffold.
Similar content being viewed by others
Introduction
Members of the formin family of cytoskeletal regulators are associated with a wide variety of essential eukaryotic cellular processes1,2. Formins vary in their biochemical functions, which include actin filament nucleation, processive association with growing filament ends, actin filament bundling, microtubule binding/stabilization, and actin filament severing (reviewed by Chesarone et al.3). These functions are usually executed by the formin homology (FH) 1 and 2 domains in the C-terminal part of the protein (Fig. 1B). The FH2 domain is conserved from yeast to humans4,5 and is a mostly α-helical homodimer with a donut-like architecture that encircles the dynamic “barbed” end of the actin filament6,7,8,9. The FH1 domain is an unstructured proline-rich region that binds profilin, an abundant actin monomer-binding protein10,11. Together these domains can modulate the growth of actin filaments12 and also displace other barbed end binding proteins13,14. In many formin isoforms, the N-terminal region contains regulatory domains that suppress the activity of the FH1 and FH2 domains15,16,17.
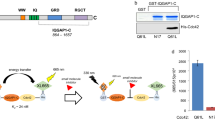
Formin constructs and purification. (A) Phylogenetic tree of formins based on DeWard et al.2 To assay diverse human formins, we selected representatives of six of the seven mammalian formin subclasses. (B) ‘FFC’ constructs, which include the FH1 and FH2 domains and all C-terminal sequences, were isolated for each formin (DID, Diaphanous Inhibitory Domain; DAD, Diaphanous Autoregulatory Domain). (C) SDS-PAGE showing the purity of each recombinant formin FFC fragment after purification by Coomassie staining (see Figure S7 for uncropped gel).
SMIFH2 (1-(3-bromophenyl)-5-(furan-2-ylmethylene)-2-thioxodihydropyrimidine-4,6(1H,5H)-dione) was discovered in an in vitro screen for compounds that inhibit the mDia FH2 domain, hence its name Small Molecule Inhibitor of FH218. SMIFH2 is assumed to be a pan-formin inhibitor based on biochemical data showing that it can inhibit yeast formins Bni1, Fus1 and Cdc12, the nematode formin CYK-1, and the mouse formin DIAPH118. More recent data show SMIFH2 interactions with plant formins including Arabidopsis formin-119, among others. However, the assumption that SMIFH2 is a pan-inhibitor has never been tested in vitro for diverse mammalian FH2 domains. To probe how formins contribute to cellular processes, both pan inhibitors as well as specific inhibitors are useful. An isoform-specific inhibitor of mouse Dia1 and Dia2 but not Dia3 has been reported, but low solubility in cell culture media has hampered its use20.
Small molecule inhibitors have been invaluable in studying cytoskeletal function. These include natural product compounds that act directly on cytoskeletal filaments21, as well as the mostly synthetic compounds that target cytoskeletal binding proteins like SMIFH2, the Arp2/3 inhibitor CK66622, and the myosin inhibitor blebbistatin and its analogs23. Due to the importance of formin-mediated cytoskeletal regulation in eukaryotes, SMIFH2 has been used in at least 324 studies of the cellular roles of formins in building both the actin and microtubule cytoskeletons (original manuscripts in a Google Scholar search for ‘SMIFH2’, May 2022).
In the context of this widespread usage, several off-target SMIFH2 interactions have been reported: one with the tumor suppressor p5324 and another with several members of the myosin superfamily25. The latter is particularly concerning, given that formins and myosins are both cytoskeletal regulators, and SMIFH2-induced effects on the cytoskeleton may be difficult to interpret in certain settings. Surprisingly, SMIFH2 more potently inhibits the actin-induced ATPase activity of Drosophila myosin 5 (IC50 ≈ 2 µM) than formin/mediated actin interactions (IC50 ≈ 10–20 µM)25. These off-target interactions may be related to the fact that SMIFH2 is classified as a Pan Assay INterference compound (or PAIN) due to the electrophilicity of its α/β-unsaturated dicarbonyl alkylidene moiety26. Both the widespread use and potential drawbacks of SMIFH2 have motivated a search for either optimized analogs of SMIFH2 or even a new generation of formin inhibitors.
In this study, we report a structure–activity study of SMIFH2 against a panel of diverse human formins, using the in vitro pyrene-actin polymerization assay27. We find that SMIFH2 is a pan-inhibitor of these human formins, and that perturbations to its structure modulate activity but not specificity.
Results and discussion
SMIFH2 was selected from a library of drug-like molecules for its ability to inhibit the mouse formin mDia1 in a pyrene-actin polymerization assay18. We used the pyrene-actin polymerization assay to characterize the inhibitory activity against a panel of human formins representing six of the seven classes of mammalian formins (Fig. 1A). These formins were expressed recombinantly as constitutively active ‘FFC’ fragments (containing FH1, FH2, and C-terminal regions; Fig. 1B) and purified using affinity chromatography (Fig. 1C).
SMIFH2 and its analogs were synthesized as described in the Experimental Procedures. In all NMR characterization experiments, it was observed that SMIFH2 was a mix of E and Z isomers, with a doubling of each proton peak due to the different environments in each isomer. In some cases, an uneven distribution of isomers was observed immediately after dissolving SMIFH2, as shown for the solvent deuterated tetrahydrofuran (THF-d8) (Fig. 2). After incubating at room temperature for 20 h, this sample equilibrated to a 1:1 mixture of isomers, as judged by the equivalent peak integrations for each pair. The shift from unequal to equal peak integrations indicates that the equilibrium constant for the isomerization process is close to 1 and that the molecule exchanges between these isomers on an experimentally relevant timescale. From NMR analysis of SMIFH2 in deuterated dimethyl sulfoxide (DMSO-d8), the initial ratio of isomers was closer to 1:1 than was observed in THF-d8. Based on this, we infer that the equilibration kinetics are faster in DMSO-d8 than in THF-d8. Regardless of solvent, we cannot say which peak of each pair corresponds to which isomer. Furthermore, it is unknown whether both isomers or just one isomer inhibits the FH2 domain.
SMIFH2 is a mix of isomers that equilibrate over time. 1H NMR spectra of the same sample were collected at two time points. The blue spectrum (bottom) corresponds to the SMIFH2 sample immediately after preparation in THF-d8. The uneven peak integrations are evident in the signals for protons A, B, C, D and E. The initial isomeric ratio was calculated as 3.5:1 by integration of major resonance at 8.25 ppm and minor resonance at 8.33 ppm for proton E. After 20 h at 25 °C (red spectrum; top), the peak integrations were similar for each isomer, reflecting equilibration to a ≈1:1 E:Z mixture (isomeric ratio 1.1:1 by integration of resonances at 8.33 ppm and 8.25 ppm).
All FFC fragments were active in pyrene-actin polymerization assays, as judged by their ability to stimulate assembly over the baseline rate of actin/profilin alone (Fig. 3, comparing red and gray curves). As reported previously, diaphanous related formins (DIAPH1 and DIAPH2) and inverted formin 2 (INF2) stimulated assembly at low nanomolar concentrations (5 nM). FMNL3, FMN2, and DAAM2 had intermediate nucleation efficiency (20–25 nM formin induced actin polymerization such that steady state was reached within 2000s), and Delphilin (25 nM) had quite weak nucleation activity, as already reported for rabbit skeletal muscle actin28.
SMIFH2 has broad activity across the human formins. For each titration, pyrene fluorescence was monitored for 2 μM actin (5% pyrene-labeled)/ 4 μM S. pombe profilin. The black dashed line shows profilin/actin alone, and the red trace corresponds to addition of formin FFC to profilin/actin (5 nM DIAPH1, DIAPH2, INF2; 20 nM FMNL3; 20 nM FMN2; 25 nM DAAM2; 25 nM Delphilin). Increasing concentration of SMIFH2 is indicated by blue traces, with the µM concentration labeled to the right of each plot. The volume of DMSO added was the same for all traces, regardless of SMIFH2 concentration.
To characterize the specificity of SMIFH2, we titrated the inhibitor against each purified formin, controlling for the volume of DMSO for all polymerization reactions (Fig. 3, as indicated by blue traces). In all cases, SMIFH2 had a concentration for 50% inhibition (IC50) in the 10–20 µM range. This is comparable to the reported value of 15 µM for mouse Dia118. Among the diaphanous formins, SMIFH2 has a more potent effect on DIAPH2 (Dia3) than on DIAPH1 (Dia1), a notable reversal of preference compared to the quinoxaline-based inhibitor reported by Higgs and coworkers (compound 2 in Gauvin et al.)20.
Several SMIFH2 analogs are known to retain or lose inhibitory activity against mDia118. We sought to expand this dataset to more extensively characterize structure–activity relationships in the SMIFH2/formin complex. All 17 analogs shown in Fig. 4 were synthesized according to the scheme described in the Experimental Procedures (Fig. 6) and characterized by proton NMR (Fig. S8). Each inhibitor was tested for potential interaction with actin in the absence of formin (Figs. S3, S4). At the highest concentrations tested, we found that some inhibitors had modest effects on the polymerization of 2 µM actin (Fig. S3). These effects were less apparent in the presence of 4 µM profilin (Fig. S4), indicating that the profilin/actin interaction may mask the inhibitor interaction or suppress polymerization of a potential actin/SMIFH2 complex. This panel of 18 inhibitors was tested against five formin FFC fragments (DIAPH1, DIAPH2, INF2, FMNL3, and FMN2), and the IC50 values were measured (Fig. 4 and Table 1). One notable trend in this dataset is that the lack of specificity, noted above for SMIFH2 (Fig. 3), holds for its analogs: no modifications made to SMIFH2 led to a selective inhibitor, at least among the five formins that were tested. In other words, active inhibitors inhibited all formins and inactive inhibitors lacked activity against all formins tested.
Structure activity data for SMIFH2 and 17 analogs tested against five human formins. Each individual data point indicates an IC50 value from a titration of inhibitor in the pyrene-actin assembly assay. The average and standard deviation for each formin/analog pair is reported in Table 1. Conditions: 2 µM actin (5% pyrene-labeled), 4 µM S. pombe profilin, formin (5 nM DIAPH1-FFC, 5 nM DIAPH2-FFC, 5 nM INF2-FFC, 20–37.5 nM FMN2-FFC, 20 nM FMNL3-FFC), inhibitor with a fixed volume of DMSO. Due to low solubility, compound 9 was dissolved in DMF. Because we cannot reliably measure IC50 values above 40 µM, those points are indicated at the > 40 µM line. Individual IC50 values are listed in Table S1.
Modifications to the thiobarbiturate core were of two types: alkylation or arylation of N3 and replacement of the thiocarbonyl with an oxygen carbonyl (atom numbering in gray in Table 1 structure). The latter substitution (5) led to a complete loss of inhibitory activity (Fig. 4 and Table 1), consistent with data reported by Kovar and coworkers (compound 3 in Rizvi, et al.18). Both methylation (2) and arylation (4) of the SMIFH2 N3 were well tolerated, and the N3 phenyl analog (4) had 3–fourfold more potent inhibitory activity than SMIFH2. Surprisingly, the ethyl-substituted analog (3) had no detectable activity. This was unexpected since the high activity of compound 4 showed that the binding pocket on the FH2 domain could accommodate substituents as large as a phenyl ring. One defining feature of PAINs is non-sensical structure–activity relationships26, and it is possible that activity data for compounds 2, 3, and 4 are confounded by the other factors apart from the free energy change associated with protein/inhibitor binding. The tolerance of SMIFH2 to methylation (2) and arylation (4) of N3 further supports a model where a carbonyl tautomer (as drawn in Table 1), not a thio enol or oxygen enol29,30, is the active form of SMIFH2. The activity of compounds 2 and 4 also indicates that the –NH group of SMIFH2 most likely does not act as a hydrogen bond donor in the protein binding pocket.
Meanwhile, several alterations to the furan-ring portion of SMIFH2 had a dramatic effect on activity. Both the pyrrole (6) and thiophene (7) analogs had no detectable inhibitory effect on any formin tested. On the other hand, halogenation of the furan ring at the C5’ position, either with Cl (15) or Br (16), was well tolerated, while methylation at that same position (17) abolished activity. This is consistent with the high activity of an analog with another halogen (I) at the 5’ position, reported by Kovar and coworkers (compound 2 in Rizvi, et al.18). We synthesized an analog with an extended linker between the furan and central rings. This compound (18) had modest activity.
SMIFH2 has a Br in the meta position of the N1 ring (R2 position in Table 1). The effect of removing this Br was explored with compound 9, which showed measurable but weak to minimal activity. Compound 9 was poorly soluble in DMSO, and had to be dissolved in DMF for assays. Aside from removing the Br altogether, we found that moving this Br to the para position (8) maintained or slightly enhanced inhibitory activity. We were interested in the effects of a methoxy group on the N1 ring, and we successfully synthesized the para-methoxy analog (10). While compound 10 had reduced activity compared to meta-Br (1) or para-Br (8) SMIFH2, substitution of the furan with either Br or Cl at the C5’ position (11 or 12, respectively) or Br at the C4’ position (13) in addition to the N1 para-methoxy phenyl group appeared to have synergistic effects. This was most striking with compound 12, the most potent inhibitor in our series of compounds. Including these two substitutions together decreased IC50 values by two–ninefold (comparing compounds 1 and 12), depending on the formin. For instance, the IC50 against INF2 was decreased from 10 ± 5 µM for SMIFH2 (1) to 2 ± 1 µM for compound 12. Unlike incorporation of a halogen at the furan C4’ or C5’ positions in the para-methoxy series (11, 12, 13), addition of a Cl at furan C5’ (14) did not have increased potency compared with para-Br alone (8).
Taken together, the activity data for SMIFH2 and the panel of 17 analogs highlight two essential components of the SMIFH2 structure: the thiocarbonyl and the furan ring. Our results also indicate that SMIFH2 derivatives maintain activity when the structure is modified at the thiobarbiturate N3 position and also at the meta and para positions on the N1-phenyl ring and the 4’ and 5’ positions of the furan ring. Despite the chemical diversity explored in this study, we did not identify an isoform-specific formin inhibitor. Such an inhibitor may exist in another region of SMIFH2 chemical space not explored here, such as ortho and/or multiple substitutions on the N1-phenyl ring or furans attached to the beta carbon at their 3’ carbon, among many other potential modifications. Alternatively, the lack of specificity among the panel of molecules in this study suggests that a more specific inhibitor may require a scaffold completely different from the thiobarbiturate core of SMIFH2 in order to distinguish among highly conserved FH2 domains. Higgs and coworkers identified a quinoxalinone-based inhibitor with specificity among the diaphanous formins20, suggesting that specific targeting among FH2 domains is achievable.
To date, no atomic-resolution structural data is available for SMIFH2 bound to the FH2 domain, hindering the rational design of structural perturbations to the inhibitor. This is in contrast to the Arp2/3 inhibitor CK66622, whose x-ray crystal structure was used as a starting point for free energy perturbation calculations to optimize inhibitor/protein interactions31. The more potent analogs reported here may be candidates for structural studies of a protein/SMIFH2 complex, particularly with the human formin DIAPH2 (ortholog of mDia3). Based on IC50 values, SMIFH2 analog 12 may have a higher affinity to DIAPH2 than any other formin/inhibitor pairs measured, although this would need to be confirmed by equilibrium experiments.
To our knowledge, it is uncertain where SMIFH2 lies in the taxonomy of covalent/noncovalent or reversible/irreversible inhibitors. Its α/β unsaturated dicarbonyl motif is likely a Michael acceptor, which would react with nucleophilic amino acids, placing SMIFH2 in the covalent category32,33,34. The furan substitution at the β carbon of SMIFH2 suggests that addition of a thiol group could be reversible35. Similar compounds to SMIFH2 with a modified thiobarbiturate core have been found to inhibit mushroom tyrosinase irreversibly36, either through covalent modification of an active site residue or induction of protein misfolding. Cellular studies with SMIFH2 have shown reversal of SMIFH2’s effects after a washout37,38,39. However, whether this is due to new protein synthesis or dissociation of the SMIFH2/formin complex is unknown.
Motivated by the wide range of inhibitory activities observed in our structure–activity study (Fig. 4 and Table 1) and the potential for covalent targeting by SMIFH2, we used computational modeling to explore differences in molecular structure and reactivity among this group of molecules. We used Gaussian density functional theory calculations at the B3LYP/6311G + + (d,p) level of theory and basis set to optimize the geometry of a subset of ten molecules, selected because of their variable activities. Figure 5A shows their optimized geometries in water for both the E and Z isomers. The optimized geometry was not sensitive to basis set or solvent (Fig. S5). Overall, the molecular shape was similar for all molecules, regardless of their inhibitory activity. Torsion angles for ‘peripheral’ rings with respect to the central thiobarbiturate ring were between 89.9° and 91.5° for the bond connecting N1 of the central ring to the equivalent of the Br-phenyl ring of SMIFH2 and between 179.5° and 180° for the bond connecting Cβ of the α/β-unsaturated thiobarbiturate to C2’ of the furan ring.
Computational analysis of SMIFH2 derivatives. (A) Optimized geometries of SMIFH2 (1) and nine analogs aligned in PyMOL based on the thiourea portion (N1-C2-N3) of each molecule. (B) Global electrophilicity plotted as a difference from SMIFH2. (C) Frontier orbital energy plotted showing the gap between the lower energy HOMO and higher energy LUMO. (D) Difference in Hirshfeld charges calculated for SMIFH2 carbons that are potential sites for nucleophilic addition. The specific carbons are indicated by a red dot in the structure exemplified for SMIFH2 (1). For panels (B–D), less active molecules are denoted with light gray bars and more active molecules are denoted with dark gray bars. All structures, energies, and charges were calculated with Gaussian (see Experimental Procedures for levels of theory and basis sets). See Figure S6 for raw electrophilicity and charge values.
We probed the degree of reactivity of SMIFH2 and these nine derivatives using a global electrophilicity index40, which we calculated from single point energies of water-solvated neutral, anionic and cationic species. We found a trend of higher electrophilicity for active inhibitors (dark bars in Fig. 5B). However, at least one inactive molecule (7), which has a thiophene in place of the furan, has an electrophilicity value slightly larger than SMIFH2 (Fig. 5B). This indicates that electrophilicity may be a determinant, but not the sole determinant, of activity. Consistent with higher electrophilicity, active inhibitors also had slightly smaller HOMO/LUMO energy gaps (Fig. 5C). Finally, we calculated Hirshfeld charges for carbons that were potential sites of nucleophilic attack, namely the carbonyl carbons and the carbon in the β position relative to the carbonyls. These charges were not correlated with activity (Fig. 5D). Overall, our computational results indicate that these SMIFH2 derivatives have similar geometries and variation in electrophilicity.
The widespread use of SMIFH2 in the decade since the foundational work of Kovar and coworkers18 underscores the demand for small molecule formin inhibitors. In addition to the search for formin-specific inhibitors, recent data showing activity of the formin inhibitor SMIFH2 on myosins25 have motivated a search for formin inhibitors with minimal off-target interactions. It will be interesting to assay the series of molecules from this study for myosin inhibition and to determine whether potency against myosins and potency against formins are correlated. Moving forward, a deeper understanding of mechanisms and specificity of formin-targeting small-molecule inhibitors is important for understanding the roles of formins in cells. We found that while many alterations disrupt SMIFH2 activity, we could increase potency by approximately fivefold. However, we failed to increase specificity, suggesting that while potency can be improved, achieving specificity among closely related FH2 domains may be hard to achieve with the SMIFH2 scaffold.
Experimental procedures
Plasmid preparation
All formin FFC gene fragments were sub-cloned into a modified pET15b vector with an N-terminal 6-histidine tag, with the exception of DIAPH1, which had a C-terminal 6-histidine tag. Several human genes were codon-optimized for E. coli and obtained as g-blocks from Integrated DNA Technologies. FFC fragments were defined with amino acid numbers: DIAPH1 (Transomic BC117257, aa 549–1262; analogous to UniProt O60610-1, aa 558–1272), DIAPH2 (NP_009293 / UniProt O60879-2, aa 553–1096), FMNL3 (NP_001354764.1 / UniProt Q8IVF7-1, aa 481-1028), FMN2 (NP_064450.3 / UniProt Q9NZ56-1, aa 1192-1722), Delphilin (NP_001138590 / UniProt A4D2P6-1, aa 744-1211), INF2 (NP_071934.3 / UniProt Q27J81-1, aa 469–1249), and DAAM2 (NP_001188356.1 / UniProt Q86T65-3, aa 486-1068). Complete DNA and amino acid sequences are included in the Supporting Information.
Protein expression and purification
Formin FFC proteins were expressed in ROSETTA-DE3 E. coli cells (Novagen) by growing the cells at 37 °C in Terrific Broth to an optical density of 0.5 –1.0 at 600 nm, reducing the temperature to 18 °C for 1 h, adding 250 μM isopropyl-beta-D-thiogalactoside (IPTG), and growing overnight (≈12–17 h) at 18 °C with shaking at 250 rpm. Cells were harvested, washed with 1 × phosphate-buffered saline (PBS), and flash frozen before storing at – 80 °C.
INF2-FFC-expressing cells were lysed in Ni–NTA lysis buffer [500 mM NaCl, 50 mM NaPi pH 7.5, 1 mM ethylenediamine tetraacetic acid (EDTA), 1 mM dithiothreitol (DTT)] supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), 4 µg/mL DNaseI (Sigma #DN25), 2000-fold diluted protease inhibitor cocktail (Sigma P8849). Cells were lysed by French Press and then centrifugd at 40,000×g for 30 min. The clarified lysate was passed over a 2-mL HisTrap column (Cytiva) and eluted with Ni–NTA lysis buffer supplemented with 500 mM imidazole. Eluted protein was gel filtered on a Superdex75 16/600 column (Cytiva) equilibrated with SEC buffer [150 mM NaCl, 10 mM HEPES pH 7, 1 mM EDTA, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP)]. Purity was assessed by SDS-PAGE, and the protein was flash frozen in liquid nitrogen and stored at – 80 °C.
DAAM2-FFC-expressing cells were lysed, centrifuged, and passed over a HisTrap column as described above for INF2. Pure fractions were pooled and buffer exchanged by PD10 column (Cytiva) into SEC buffer before dialyzing with 1:1 glycerol:SEC buffer overnight. Aliquots were flash frozen and stored at – 80 °C.
DIAPH2-FFC expressing cells were lysed by French Press in Ni–NTA lysis buffer supplemented with PMSF, DNaseI and protease inhibitor cocktail and centrifuged, as described for INF2. Clarified lysates were nutated for 1 h at 4 °C with Ni–NTA resin (Thermo Sci). The resin was washed with 25 column volumes (CV) of Ni–NTA lysis buffer and 25 CV of Ni–NTA lysis buffer supplemented with 10 mM imidazole, before eluting with lysis buffer supplemented with 500 mM imidazole. Eluted protein was buffer exchanged by PD10 column into S-buffer (10 mM NaCl, 10 mM PIPES pH 6.5, 1 mM EDTA, 1 mM DTT). The protein was loaded onto a 1-mL SP-FF cation exchange column (Cytiva) and eluted with a gradient from 10 to 500 mM NaCl over 30 CV. Selected fractions were exchanged into SEC buffer by PD10 column and then dialyzed into 1:1 glycerol:SEC buffer overnight. Aliquots were flash frozen and stored at –80 °C. FMN2-FFC-expressing cells were lysed in a modified pH 6.5 Ni–NTA lysis buffer. The rest of the purification protocol was identical to that described for DIAPH2-FFC.
FMNL3-FFC-expressing cells were lysed in TALON extraction buffer (300 mM NaCl, 50 mM NaPi pH 8.0, 1 mM β-mercaptoethanol (βME)) supplemented with 1 mM PMSF and 4 µg/mL DNaseI. After lysis and centrifugation as described above for INF2-FFC, the supernatant was nutated with 1 mL TALON resin (Takara) for 30 min at 4 °C. The resin was washed with 25 CV TALON extraction buffer and then 25 CV TALON wash buffer (300 mM NaCl, 50 mM NaPi pH 7, 1 mM βME) before eluting with TALON wash buffer supplemented with 200 mM imidazole. The eluted protein was dialyzed against 50 mM NaPi pH 7, 50 mM NaCl, 1 mM DTT and loaded onto an SP-FF cation exchange column and eluted with a gradient from 50 to 500 mM NaCl over 30 CV. Fractions were dialyzed against S-buffer, loaded on an SP-FF column, and eluted with a step gradient from 10 mM NaCl to 500 mM NaCl. Eluted fractions were loaded onto a Superdex200 10/300 column (Cytiva) equilibrated with SEC buffer. Pure fractions were flash frozen in liquid nitrogen and stored at – 80 °C. FMNL3 was also purified in a protocol that skipped the first SP-FF column, and there was no difference in activity or purity.
DIAPH1-FFC-expressing cells were lysed by probe sonicator in TALON extraction buffer supplemented with PMSF and DNaseI. The formin was purified using TALON resin as described above for FMNL3-FFC. Eluted fractions were dialyzed against Q-buffer (10 mM Tris pH 8, 1 mM DTT) overnight with 100 units of thrombin to cleave the C-terminal histidine tag. The cleaved protein was loaded onto a monoQ anion exchange column (Cytiva) and eluted with a gradient of 0 to 500 mM KCl over 30 CV. Semi-pure fractions were buffer exchanged into TALON extraction buffer using a PD10 column and incubated with TALON resin to remove uncleaved protein. The fraction that did not bind TALON was collected and dialyzed against Q-buffer and then 1:1 glycerol:Q-buffer. This results in a version of DIAPH1-FFC that is truncated by ~ 10–20 residues on the C-terminus as assessed by MALDI mass spectrometry (data not shown).
Delphilin-FFC was purified as described for the human FFC isoform28. Profilin from Schizosaccharomyces pombe was expressed in a pET plasmid in BL21-DE3* cells and was purified as described41. We chose this isoform of profilin since, compared to other profilins, its interaction with actin is less sensitive to actin modification at Cys73442. Actin was purified from rabbit skeletal muscle and labeled with pyrene-iodoacetamide as described42. For pyrene-labeled actin, the pyrene concentration was calculated using an extinction coefficient of 21,978 M−1 cm−1 at 344 nm, and the actin concentration was calculated using the correction factor of 0.127 at 290 nm ([actin] = (A290 − 0.127 × A344) × 38 μM). Extinction coefficients for formins at 280 nm were calculated using Expasy ProtParam43: DIAPH1 (22,920 M−1 cm−1), DIAPH2 (25,900 M−1 cm−1), FMNL3 (28,420 M−1 cm−1), FMN2 (36,900 M−1 cm−1), Delphilin (25,440 M−1 cm−1), INF2 (47,440 M−1 cm−1), and DAAM2 (29,910 M−1 cm−1).
SMIFH2 synthesis and characterization
For the preparation of SMIFH2 and its analogs (Fig. 6), N-aryl-substituted thioureas (A, X = S, R’ = H) were prepared by treatment of the corresponding anilines with ammonium thiocyanate in aqueous acid. Potassium cyanate gave the urea intermediate (A, X = O, R’ = H) en route to analog 544. Use of the corresponding alkyl- or phenyl isothiocyanate provided N3-substituted derivatives (A, X = S, R’ = Me, Et, Ph). Reaction of the (thio)ureas A with diethyl malonate and sodium ethoxide in ethanol solution29 formed the thiobarbituric acid core of intermediates B. The synthesis of SMIFH2 and analogs (C) was completed by Knoevenagel condensation with a selection of aldehydes45,46, either under standard reflux conditions or using a microwave reactor. SMIFH2 and its analogs formed as mixtures of E/Z isomers and were characterized by 1H NMR and, in selected cases, 13C NMR and/or HRMS. Further experimental details and characterization data are included in the Supporting Information.
Pyrene-actin assays
Assays were carried out as described47. Rabbit skeletal muscle actin (5% pyrene labeled) was incubated for 2 min at 25 °C with 200 μM EGTA and 50 μM MgCl2 to convert Ca-actin to Mg-actin. When included in the experiment, a 2:1 molar ratio of profilin was incubated with actin for 2 min at 25 °C before conversion to Mg-actin. Polymerization was initiated by adding polymerization buffer (KMEH, final concentration: 10 mM HEPES, pH 7.0, 1 mM EGTA, 50 mM KCl, 1 mM MgCl2). Prior to mixing KMEH with actin, formins and then the inhibitor in DMSO or DMF was added at ≤ 1% of the total volume. Finally, this mixture was rapidly mixed with Mg-actin, with approximately 10–20 s dead time prior to measurement. Fluorescence was monitored every 15 s using a TECAN F200 Pro with λex = 360 ± 17 nm and λem = 415 ± 10 nm. The same volume of organic solvent (DMSO or DMF) was added to each well regardless of inhibitor concentration.
Computational analysis
SMIFH2 isomers and their chemical analogs were constructed using the program Avogadro48. All quantum mechanical calculations were carried out using Gaussian 16 (versions A-2016 and C-2019)49. Geometry optimizations, single point energy, and Hirshfeld population calculations were performed with the unrestricted (U) B3LYP functional with the 6–311 + G(d,p) basis set50,51,52,53,54,55,56,57,58,59,60,61,62. For geometry optimizations, the opt keyword was set to the ‘tight’ option to ensure adequate convergence. The initial optimization for all compounds was performed in vacuum and used further for solvated calculations reported in this study. Single point energy calculations, for a global electrophilicity analysis, at the neutral, anionic, and cationic states were performed with the unrestricted (U) PW6B95 functional supplemented with Grimme et al.’s D3 empirical diffuse function and the aug-cc-pVTZ basis set63,64,65,66,67. SCRF polarizable continuum model (PCM) was used to model an implicit water as the solvent. Optimized geometries were visualized using PyMOL68.
To calculate global electrophilicity (ω), we used the convention of Parr et al. ω = µ2/(2η) where µ and η represent the electronic chemical potential and the chemical hardness, respectively40,69,70,71,72. Ionization energy and electron affinity were used to calculate µ and η, as defined by De Vleeschouwer et al.73 We used the (U) PW6B95-D3 functional and aug-cc-pVTZ basis set as described above to determine the global electrophilicity of SMIFH2 and its analogs more accurately74.
Data availability
References
Goode, B. L. & Eck, M. J. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593–627 (2007).
Deward, A. D., Eisenmann, K. M., Matheson, S. F. & Alberts, A. S. The role of formins in human disease. Biochim. Biophys. Acta 1803, 226–233 (2010).
Chesarone, M. A., Dupage, A. G. & Goode, B. L. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 11, 62–74 (2010).
Higgs, H. N. & Peterson, K. J. Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell 16, 1–13 (2005).
Pruyne, D. Revisiting the phylogeny of the animal formins: two new subtypes, relationships with multiple wing hairs proteins, and a lost human formin. PLoS ONE 11, e0164067 (2016).
Xu, Y. et al. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell 116, 711–723 (2004).
Otomo, T. et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 433, 488–494 (2005).
Lu, J. et al. Structure of the FH2 domain of Daam1: Implications for formin regulation of actin assembly. J. Mol. Biol. 369, 1258–1269 (2007).
Thompson, M. E., Heimsath, E. G., Gauvin, T. J., Higgs, H. N. & Kull, F. J. FMNL3 FH2–actin structure gives insight into formin-mediated actin nucleation and elongation. Nat. Struct. Mol. Biol. 20, 111–118 (2012).
Courtemanche, N. & Pollard, T. D. Determinants of Formin homology 1 (FH1) domain function in actin filament elongation by formins. J. Biol. Chem. 287, 7812–7820 (2012).
Horan, B. G., Zerze, G. H., Kim, Y. C., Vavylonis, D. & Mittal, J. Computational modeling highlights the role of the disordered Formin homology 1 domain in profilin-actin transfer. FEBS Lett. 592, 1804–1816 (2018).
Kovar, D. R., Harris, E. S., Mahaffy, R., Higgs, H. N. & Pollard, T. D. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–435 (2006).
Zigmond, S. H. et al. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 13, 1820–1823 (2003).
Bombardier, J. P. et al. Single-molecule visualization of a formin-capping protein ‘decision complex’ at the actin filament barbed end. Nat. Commun. 6, 8707 (2015).
Li, F. & Higgs, H. N. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13, 1335–1340 (2003).
Liu, W. et al. Mechanism of activation of the Formin protein Daam1. Proc. Natl. Acad. Sci. U.S.A. 105, 210–215 (2008).
Bor, B., Vizcarra, C. L., Phillips, M. L. & Quinlan, M. E. Autoinhibition of the formin Cappuccino in the absence of canonical autoinhibitory domains. Mol. Biol. Cell 23, 3801–3813 (2012).
Rizvi, S. A. et al. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16, 1158–1168 (2009).
Cao, L., Henty-Ridilla, J. L., Blanchoin, L. & Staiger, C. J. Profilin-dependent nucleation and assembly of actin filaments controls cell elongation in Arabidopsis. Plant Physiol. 170, 220–233 (2016).
Gauvin, T. J., Fukui, J., Peterson, J. R. & Higgs, H. N. Isoform-selective chemical inhibition of mDia-mediated actin assembly. Biochemistry 48, 9327–9329 (2009).
Risinger, A. L. & Du, L. Targeting and extending the eukaryotic druggable genome with natural products: Cytoskeletal targets of natural products. Nat. Prod. Rep. 37, 634–652 (2020).
Nolen, B. J. et al. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031–1034 (2009).
Rauscher, A. Á., Gyimesi, M., Kovács, M. & Málnási-Csizmadia, A. Targeting myosin by blebbistatin derivatives: Optimization and pharmacological potential. Trends Biochem. Sci. 43, 700–713 (2018).
Isogai, T., van der Kammen, R. & Innocenti, M. SMIFH2 has effects on Formins and p53 that perturb the cell cytoskeleton. Sci. Rep. 5, 9802 (2015).
Nishimura, Y. et al. The formin inhibitor SMIFH2 inhibits members of the myosin superfamily. J. Cell Sci. 134, 2 (2021).
Baell, J. B. Observations on screening-based research and some concerning trends in the literature. Fut. Med. Chem. 2, 1529–1546 (2010).
Cooper, J. A., Walker, S. B. & Pollard, T. D. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell. Motil. 4, 253–262 (1983).
Silkworth, W. T. et al. The neuron-specific formin Delphilin nucleates nonmuscle actin but does not enhance elongation. Mol. Biol. Cell 29, 610–621 (2018).
Wang, G. et al. Experimental and theoretical investigations on the tautomerism of 1-phenyl-2-thiobarbituric acid and its methylation reaction. J. Mol. Struct. 1036, 372–379 (2013).
Mahmudov, K. T. et al. Barbituric acids as a useful tool for the construction of coordination and supramolecular compounds. Coord. Chem. Rev. 265, 1–37 (2014).
Baggett, A. W. et al. Structural characterization and computer-aided optimization of a small-molecule inhibitor of the Arp2/3 complex, a key regulator of the actin cytoskeleton. ChemMedChem 7, 1286–1294 (2012).
Schwöbel, J. A. H. et al. Measurement and estimation of electrophilic reactivity for predictive toxicology. Chem. Rev. 111, 2562–2596 (2011).
Jackson, P. A., Widen, J. C., Harki, D. A. & Brummond, K. M. Covalent modifiers: a chemical perspective on the reactivity of α, β-unsaturated carbonyls with thiols via hetero-Michael addition reactions. J. Med. Chem. 60, 839–885 (2017).
Tuley, A. & Fast, W. The taxonomy of covalent inhibitors. Biochemistry 57, 3326–3337 (2018).
Krenske, E. H., Petter, R. C. & Houk, K. N. Kinetics and thermodynamics of reversible thiol additions to mono- and diactivated Michael acceptors: implications for the design of drugs that bind covalently to cysteines. J. Org. Chem. 81, 11726–11733 (2016).
Yan, Q. et al. Inhibitory effects of 5-benzylidene barbiturate derivatives on mushroom tyrosinase and their antibacterial activities. Eur. J. Med. Chem. 44, 4235–4243 (2009).
Hu, S. et al. Long-range self-organization of cytoskeletal myosin II filament stacks. Nat. Cell Biol. 19, 133–141 (2017).
Qu, X. et al. Stabilization of dynamic microtubules by mDia1 drives Tau-dependent Aβ1-42 synaptotoxicity. J. Cell Biol. 216, 3161–3178 (2017).
Fenix, A. M. et al. Muscle-specific stress fibers give rise to sarcomeres in cardiomyocytes. Elife 7, e42144 (2018).
Parr, R. G., Szentpály, L. V. & Liu, S. Electrophilicity index. J. Am. Chem. Soc. 121, 1922–1924 (1999).
Bradley, A. O., Vizcarra, C. L., Bailey, H. M. & Quinlan, M. E. Spire stimulates nucleation by Cappuccino and binds both ends of actin filaments. Mol. Biol. Cell 31, 273–286 (2020).
Lakha, R. et al. Variable autoinhibition among deafness-associated variants of Diaphanous 1 (DIAPH1). Biochemistry 60, 2320–2329 (2021).
Gasteiger, E. et al. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook (ed. Walker, J. M.) 571–607 (Humana Press, 2005). https://doi.org/10.1385/1-59259-890-0:571.
Saeed, S., Rashid, N., Jones, P. G., Ali, M. & Hussain, R. Synthesis, characterization and biological evaluation of some thiourea derivatives bearing benzothiazole moiety as potential antimicrobial and anticancer agents. Eur. J. Med. Chem. 45, 1323–1331 (2010).
Bazin, M., Harriman, G., Kuhn, C. & Luly, J. Barbiturates as integrin antagonists and their use for treating inflammatory diseases. US20040214845A1 (2004).
Liou, H., Liou, M.-L. & Ouk, S. C-Rel inhibitors and uses thereof. US9873674B2 (2018).
Zuchero, J. B. In vitro actin assembly assays and purification from Acanthamoeba. Methods Mol. Biol. 370, 213–226 (2007).
Hanwell, M. D. et al. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics 4, 17 (2012).
Frisch, M. J. et al. Gaussian 16 Rev. C.01. (2016).
Wachters, A. J. H. Gaussian basis set for molecular wavefunctions containing third-row atoms. J. Chem. Phys. 52, 1033–1036 (1970).
Hay, P. J. Gaussian basis sets for molecular calculations. The representation of 3d orbitals in transition-metal atoms. J. Chem. Phys. 66, 4377–4384 (1977).
Hirshfeld, F. L. Bonded-atom fragments for describing molecular charge densities. Theoret. Chim. Acta 44, 129–138 (1977).
Krishnan, R., Binkley, J. S., Seeger, R. & Pople, J. A. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys. 72, 650–654 (1980).
McLean, A. D. & Chandler, G. S. Contracted Gaussian basis sets for molecular calculations. I. Second row atoms, Z=11–18. J. Chem. Phys. 72, 5639–5648 (1980).
Clark, T., Chandrasekhar, J., Spitznagel, G. W. & Schleyer, P. V. R. Efficient diffuse function-augmented basis sets for anion calculations. III. The 3–21+G basis set for first-row elements, Li–F. J. Comp. Chem. 4, 294–301 (1983).
Frisch, M. J., Pople, J. A. & Binkley, J. S. Self-consistent molecular orbital methods 25. Supplementary functions for Gaussian basis sets. J. Chem. Phys. 80, 3265–3269 (1984).
Ritchie, J. P. Electron density distribution analysis for nitromethane, nitromethide, and nitramide. J. Am. Chem. Soc. 107, 1829–1837 (1985).
Ritchie, J. P. & Bachrach, S. M. Some methods and applications of electron density distribution analysis. J. Comp. Chem. 8, 499–509 (1987).
Binning, R. C. Jr. & Curtiss, L. A. Compact contracted basis sets for third-row atoms: Ga–Kr. J. Comp. Chem. 11, 1206–1216 (1990).
McGrath, M. P. & Radom, L. Extension of Gaussian-1 (G1) theory to bromine-containing molecules. J. Chem. Phys. 94, 511–516 (1991).
Becke, A. D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652 (1993).
Curtiss, L. A. et al. Extension of Gaussian-2 theory to molecules containing third-row atoms Ga–Kr. J. Chem. Phys. 103, 6104–6113 (1995).
Kendall, R. A., Dunning, T. H. & Harrison, R. J. Electron affinities of the first-row atoms revisited. Systematic basis sets and wave functions. J. Chem. Phys. 96, 6796–6806 (1992).
Woon, D. E. & Dunning, T. H. Gaussian basis sets for use in correlated molecular calculations. III. The atoms aluminum through argon. J. Chem. Phys. 98, 1358–1371 (1993).
Davidson, E. R. Comment on “Comment on Dunning’s correlation-consistent basis sets”. Chem. Phys. Lett. 260, 514–518 (1996).
Zhao, Y. & Truhlar, D. G. Design of density functionals that are broadly accurate for thermochemistry, thermochemical kinetics, and nonbonded interactions. J. Phys. Chem. A 109, 5656–5667 (2005).
Grimme, S., Antony, J., Ehrlich, S. & Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 132, 154104 (2010).
Schrodinger. The PyMOL Molecular Graphics System, Version 1.8. (2015).
Parr, R. G., Donnelly, R. A., Levy, M. & Palke, W. E. Electronegativity: the density functional viewpoint. J. Chem. Phys. 68, 3801–3807 (1978).
Parr, R. G. & Pearson, R. G. Absolute hardness: Companion parameter to absolute electronegativity. J. Am. Chem. Soc. 105, 7512–7516 (1983).
Geerlings, P., De Proft, F. & Langenaeker, W. Conceptual density functional theory. Chem. Rev. 103, 1793–1874 (2003).
Gunasekara, T. et al. TEMPO-mediated catalysis of the sterically hindered hydrogen atom transfer reaction between (C5Ph5)Cr(CO)3H and a trityl radical. J. Am. Chem. Soc. 141, 1882–1886 (2019).
De Vleeschouwer, F., Van Speybroeck, V., Waroquier, M., Geerlings, P. & De Proft, F. Electrophilicity and nucleophilicity index for radicals. Org. Lett. 9, 2721–2724 (2007).
Goerigk, L. et al. A look at the density functional theory zoo with the advanced GMTKN55 database for general main group thermochemistry, kinetics and noncovalent interactions. Phys. Chem. Chem. Phys. 19, 32184–32215 (2017).
Acknowledgements
The authors acknowledge the work of the Advanced Chemical Synthesis Laboratory students (Amber Scharnow, Anna Yusov, Jacqueline Chou, Karen Montero, Karol Francisco, Kelsey Lynch, Kiana Harris, Lina Davidson, Maria Paley, Tahrima Jalil, Aisha Hasan, Amanda Sosnowski, Andromeda Urquilla, Choi Mak, Emeline Nguyenduy, Isabel Klein, Jenny Lam, Rachel Dziatko, Ataa Amponsah, Elizabeth Witta, Erica Christensen, Hannah Wentz, Irene Golden, Isabelle Rocroi, Rafaela Brinn, Alaina Hartnett, Allison Forsberg, Anna Hurdle, Emily Latif, Genevieve Nemeth, Janine Sempel, Jessica Glynn, Lucy Zorzano, Shoshana Williams, Tasneem Elkoush, Wendy Xie, Dominique Macaluso, Emily Miura-Stempel, Erika Amemiya, Isabel Hernandez-Rodriguez, Jennifer Niola, Juliet Lee, Kalina Ko, Maya Hoffman, Michelle Lin, Natasha Reich). Dina Merrer, Marisa Buzzeo, and Dalibor Sames for helpful discussion, Rabina Lakha for actin purification, Grace Nickel for help with molecular biology.
Funding
This work was supported by the Research Corporation for Scientific Advancement (Cottrell award #25929 to CLV), Barnard College, and XSEDE (allocation # MCB200054 to CLV). The NMR spectrometer was supported by the NSF Division of Chemistry (MRI award #1827936).
Author information
Authors and Affiliations
Contributions
M.O., M.L., D.N., J.G., K.S., V.H., A.K., C.H., S.Q.L., J.M.K., J.J.K., C.M.R., and C.L.V. acquired and analyzed data. A.O. and J.M.K. carried out computational analysis. M.O., S.Q.L., J.M.K., F.B., J.J.K., C.M.R., and C.L.V. designed experiments. A.O., D.N., J.G., F.B., C.M.R., and C.L.V. prepared the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Orman, M., Landis, M., Oza, A. et al. Alterations to the broad-spectrum formin inhibitor SMIFH2 modulate potency but not specificity. Sci Rep 12, 13520 (2022). https://doi.org/10.1038/s41598-022-17685-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-17685-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.