Abstract
t(6;9)(p22;q34.1)/DEK-NUP214 is a recurrent genetic abnormality that occurs in 1–2% of patients with acute myeloid leukemia (AML), and rarely in myelodysplastic syndrome (MDS). It has been suggested by others that all myeloid neoplasms with t(6;9)/DEK-NUP214 may be considered as AML, even when blast count is <20%. In this study, we compared the clinicopathologic features of 107 patients with myeloid neoplasms harboring t(6;9)/DEK-NUP214: 33 MDS and 74 AML. Compared with patients with AML, patients with MDS were older (p = 0.10), had a lower white blood cell count (p = 0.0017), a lower blast count in the peripheral blood (p < 0.0001) and bone marrow (p < 0.0001), a higher platelet count (p = 0.022), and a lower frequency of FLT3-ITD mutation (p = 0.01). In addition, basophilia was not a common feature in the patients of this cohort. Although there was no difference in overall survival between MDS and AML patients (p = 0.18) in the entire cohort, the survival curves did show a trend toward favorable survival in MDS patients. Multivariate analyses showed that initial diagnosis of MDS vs. AML and allogeneic hematopoietic stem cell transplantation were prognostic factors for survival of patients with t(6;9)/DEK-NUP214 (p = 0.008 and p < 0.0001, respectively). Our data suggest that MDS with t(6;9)/DEK-NUP214 is prognostically not equivalent to AML with t(6;9)/DEK-NUP214. These data also show that stem cell transplantation greatly improves the survival of MDS and AML patients with myeloid neoplasms associated with t(6;9)/DEK-NUP214.
Similar content being viewed by others
Introduction
t(6;9)(p22;q34.1)/DEK-NUP214 is a recurrent genetic abnormality that occurs in 1–2% of patients with acute myeloid leukemia (AML) [1, 2]. AML associated with t(6;9)/DEK-NUP214 has been recognized as a distinct entity in the World Health Organization (WHO) classification since 2008 [3]. This translocation in AML was first identified by Rowley and Potter in 1976 [4], and was shown subsequently to involve DEK on chromosome 6p22.3 and NUP214 (formerly known as CAN) on chromosome 9q34.1, leading to formation of a DEK-NUP214 fusion gene. Consistent with its association with AML, increased expression of the fusion protein has been reported to be restricted to cells of myeloid lineage [5, 6].
AML associated with t(6;9)/DEK-NUP214 has morphological features that overlap with AML with maturation or acute myelomonocytic leukemia [1, 7]. Frequently, these neoplasms show evidence of myelodysplasia and in some cases, basophilia, Auer rods, and ring sideroblasts [1, 7,8,9,10]. These neoplasms also commonly harbor an internal tandem duplication (ITD) of FLT3 [1, 7, 9,10,11,12]. Others have reported that adults and children with AML associated with t(6;9)/DEK-NUP214 have a poor prognosis [1, 2, 8, 9, 11, 13], and data suggest that allogeneic hematopoietic stem cell transplantation (HSCT) may improve survival [1, 12].
Rarely, the t(6;9)/DEK-NUP214 has been reported in cases of myelodysplastic syndrome (MDS) [8, 10, 14,15,16,17,18]. These patients have a high risk of progression to AML [8, 10, 14, 18,19,20]. In a study of 62 pediatric patients including 8 patients with MDS and 54 with AML associated with t(6;9)/DEK-NUP214, Sandahl et al. found similar clinical and morphological characteristics between t(6;9)/DEK-NUP214-positive MDS and AML, suggesting that MDS with t(6;9)/DEK-NUP214 may be better considered as AML regardless of the blast percentage, analogous to myeloid neoplasms associated with t(8;21)(q22;q22.1)/RUNX1-RUNX1T1, and inv(16)(p13.1q22) or t(16;16)(p13.1;q22)/CBFB-MYH11 [9].
MDS associated with t(6;9)/DEK-NUP214 is rare and only a few small series have been reported. No large studies have compared patients with MDS with t(6;9)/DEK-NUP214 vs. AML with t(6;9)/DEK-NUP214. In this multicenter study of 107 patients, we compared the clinicopathologic features of patients with MDS vs. AML harboring t(6;9)/DEK-NUP214. Our aim was to address the clinical and prognostic impact of t(6;9)/DEK-NUP214 in MDS, and more specifically, address this question: should MDS associated with t(6;9)/DEK-NUP214 be classified as AML, even with a blast percentage of <20%?
Materials and methods
Patient selection and data collection
Cases of myeloid neoplasms diagnosed at The University of Texas MD Anderson Cancer Center since 1997 were reviewed for the presence of t(6;9)/DEK-NUP214 detected by conventional chromosomal analysis, fluorescence in situ hybridization (FISH), and/or molecular testing, yielding 16 MDS and 50 AML cases. An additional 41 cases (17 MDS and 24 AML) were contributed from nine collaborating institutions where the contributors prioritized identification of MDS cases. Clinical information and follow-up data were obtained from the electronic medical records, including age, gender, history of cytotoxic therapy before the emergence of t(6;9)/DEK-NUP214, initial pathological diagnosis, immunophenotyping, interval from initial diagnosis of MDS to progression to AML, therapy, clinical response, status of last follow-up, and overall survival (OS). The types of treatment included standard 7 + 3 chemotherapy (cytarabine and anthracycline), usually with additional chemotherapies including etoposide, hypomethylating agents (HMA) including azacitidine and decitabine, targeted therapies including FLT3 inhibitors, and HSCT. Laboratory findings at initial diagnosis of MDS or AML were recorded, including complete blood count and differential counts. This study was approved by the Institutional Review Boards of all participating institutions.
Morphologic assessment
Wright–Giemsa-stained peripheral blood (PB) and bone marrow (BM) aspirate smears and hematoxylin-eosin-stained BM clot and core biopsy sections were reviewed. These specimens were assessed for morphologic dysplasia of all lineages. The assessment of dysplasia followed criteria described previously [7]. A total of 200 leukocytes in PB and 500 cells in BM were counted to determine cell differentiation including blast and basophil percentages. Evidence of progression to AML in patients with MDS was also evaluated, defined as the occurrence of ≥20% blasts in BM or PB after the initial diagnosis of MDS.
Chromosomal, FISH analyses, and acute leukemia translocation panel
Conventional chromosomal analysis was performed on G-banded metaphases prepared from unstimulated 24- and 48-h BM aspirate cultures using standard GTG banding. Twenty metaphases were analyzed, and the results were reported using the 2016 International System for Human Cytogenetics Nomenclature. A complex karyotype was defined as ≥3 chromosomal abnormalities. FISH analysis for DEK-NUP214 was performed on cultured cells with DEK-NUP214 dual-color dual fusion probes (Abbott Molecular, Des Plaines, IL, USA) according to the manufacturer’s instructions. Two hundred interphase nuclei were analyzed. The presence of one red, one green, and two yellow (fusion) signals was considered as the typical signal pattern for DEK-NUP214 fusion. Nanofluidics-based qualitative multiparametric reverse-transcriptase PCR was performed for the detection of a panel of fusion transcripts associated with acute leukemia including t(6;9)/DEK-NUP214 on MDS or AML patients at MDACC, with an analytical sensitivity of 0.1–0.01% [21].
Flow cytometric analysis
Flow cytometry immunophenotypic analysis was performed on fresh BM aspirate specimens collected in tubes containing ethylenediaminetetraacetic acid as part of routine clinical service in the clinical flow cytometry laboratories of each institution. Samples were examined with antibody panels designed to assess acute leukemia, and the antibodies used mainly to evaluate myeloblasts were as follows: CD2, surface and cytoplasmic CD3, CD4, CD5, CD7, CD9, CD10, CD13, CD14, CD15, CD19, CD33, CD34, CD36, CD38, CD41, CD56, CD64, CD117, CD123, HLA-DR, MPO, and TdT. At MDACC, the acquisition of events was performed on FACSCalibur instruments (BD Biosciences, San Jose, CA, USA), in which standardization was maintained using CS&T Beads with emphasis on comparable cross platform performance. Data were analyzed using FCS Express (De Novo Software, Glendale, CA).
Mutation analysis
Different panels designed to detect genes commonly mutated in myeloid neoplasms were performed on fresh-frozen BM DNA samples over different time intervals and at different institutions using targeted next-generation sequencing (NGS) methods (Supplementary Materials and Methods: NGS panels). A 28- or 81-gene panel at MDACC was performed using the same methods as described previously [22, 23]. A 30- or 49-gene panel was used for cases contributed by Memorial Sloan Kettering Cancer Center (New York, NY, USA), and the ARUP Myeloid Malignancies Mutation Panel was used by University of Nebraska Medical Center (Omaha, NE, USA). Certain genes were tested separately with or without NGS panels. FLT3-ITD, FLT3 D835, and NPM1 (exon 12, codons 956-971) mutations were assessed by PCR followed by capillary electrophoresis on a genetic analyzer (Prism ABI 3130, Applied Biosystems, Foster City, CA, USA) as described previously [24]. NRAS (codons 12, 13, 61), KRAS (codons 12, 13, 61), IDH1 (exon 4, codons 87–138), IDH2 (exon 4), KIT (exons 8 and 17), DNMT3A (exon 23), CEBPA, and JAK2 (codon 617) mutations were assessed by PCR followed by Sanger sequencing with a sensitivity of 10–20% as described previously [25, 26].
Statistical analysis
Clinical and laboratory variables were compared between patients with MDS vs. AML using chi-square or Fisher’s exact tests for categorical variables and unpaired t-test or Mann–Whitney U tests for numeric variables. The interval from MDS to AML was calculated as the time from the initial diagnosis of MDS to the diagnosis of AML progression. OS was calculated from the initial diagnosis of MDS or AML to the date of expiration or the last known alive date. Survival distributions were analyzed with the Kaplan–Meier method, with differences compared by the log-rank test. To compare OS of patients with AML associated with t(6;9)/DEK-NUP214 in our cohort to a more general AML population, a public database from the TCGA AML study was used and only patients equal to or younger than 55 years were included in the survival analysis to achieve a patient group of similar median age to our study cohort [27].
To perform multivariate analysis, a univariate cox model was run for all variables at initial detection of t(6;9)/DEK-NUP214, including initial diagnosis (MDS vs. AML), age, white blood cell count, hemoglobin, platelet count, erythroid dysplasia, granulocytic dysplasia, megakaryocytic dysplasia, multilineage dysplasia, complex karyotype, isolated t(6;9), FLT3-ITD mutation, 7 + 3 chemotherapy, HMA, FLT3 inhibitor treatment, and HSCT status. Variables significant at some level (p ≤ 0.20) were included in the initial multivariate model. Then using stepwise regression procedure, the final main-effect model with a significant level (p < 0.05) was performed after backward selection to eliminate non-significant variables at some level (p ≥ 0.10) and forward selection to add variables with significant level p ≤ 0.10. Statistical analyses were performed using GraphPad Prism 8 (La Jolla, CA, USA) and R statistical software (version 4.0.2, R core team).
Results
Baseline clinical characteristics
The study cohort included 107 patients with myeloid neoplasms harboring t(6;9)/DEK-NUP214: 33 patients with MDS and 74 with AML. There were 55 men and 52 women with a median age of 43 years (range, 10–82) at initial diagnosis. Patients with MDS were older than those with AML (median, 51 vs. 38 years) but this difference was not significant (p = 0.10). Similarly, neither patients with MDS nor patients with AML showed a gender predilection. The clinical, morphologic, and genetic characteristics of the cohort are summarized in Table 1.
In all 101 patients with known time of t(6;9)/DEK-NUP214 emergence, the translocation was detected at time of initial diagnosis of MDS or AML. In the remaining six patients, the time of emergence of t(6;9)/DEK-NUP214 was unknown. In these patients, conventional chromosomal analysis, FISH, and molecular testing for DEK-NUP214 were not performed at initial diagnosis and t(6;9)/DEK-NUP214 was detected at follow-up. Four of these patients were diagnosed initially with MDS, which progressed to AML during follow-up; t(6;9)/DEK-NUP214 was identified 6–13 months after AML progression. However, an earlier karyotype was not available and the possibility of t(6;9)/DEK-NUP214 occurring as a secondary event cannot be excluded.
Laboratory findings
Patients with MDS associated with t(6;9)/DEK-NUP214 presented with a lower white blood cell count (median: 3.2 vs. 15.3 × 109/L, p = 0.0017) and a higher platelet count (median: 85 vs. 52 × 109/L, p = 0.022) than patients with AML (Table 1). There was no significant difference in the hemoglobin level (p = 0.22) and basophil percentages in the PB (p = 0.61) or BM (p = 0.1) between patients with MDS and AML. Of the 96 patients with the PB or BM differential counts available, 7 (7%) had basophils over 2% in the PB and/or BM. In the PB, 66/86 (77%) patients had no basophils in the differential count, including 48/61 (79%) with AML and 18/25 (72%) MDS. In the BM, 68/94 (72%) patients had no basophils, including 46/68 (68%) with AML and 22/26 (85%) MDS. As expected, patients with MDS presented with a lower blast count in the PB (median, 0% vs. 12%, p < 0.0001) and BM (median, 11% vs. 56%, p < 0.0001) than those with AML.
Morphologic findings
The classification of the 33 MDS cases using the WHO system was as follows: 15 MDS with excess blasts-2 (MDS-EB2), 10 MDS-EB1, 4 MDS with multilineage dysplasia, 1 atypical chronic myeloid leukemia, BCR-ABL1-negative, and 3 MDS of unknown sub-classification. By morphology only, the 74 AML cases were classified as follows: 40 (54%) AML with maturation, 26 (35%) acute myelomonocytic leukemia, 3 (4%) acute monocytic leukemia, 2 (3%) AML without maturation, and 3 (4%) without morphologic classification available.
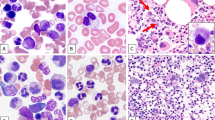
A variable degree of multilineage dysplasia was seen in 69 of 99 (70%) cases with BM materials available for morphologic evaluation (Fig. 1), including dyserythropoiesis in 75 (75%), dysgranulopoiesis in 63 (64%), and dysmegakaryopoiesis in 55 (56%). MDS with t(6;9)/DEK-NUP214 showed a higher frequency of multilineage dysplasia than AML with t(6;9)/DEK-NUP214 (27/30, 90% vs. 42/69, 61%, p = 0.004). This difference seems attributable to a higher frequency of dysplasia in the megakaryocytic lineage in MDS with t(6;9)/DEK-NUP214 (25/30, 83% vs. 30/69, 43%, p = 0.0003). Sixty cases were evaluated specifically for Auer rods; in 11 (18%) cases Auer rods were present in blasts.
A Bone marrow from a patient with MDS-EB2 with t(6;9) and mutations of NRAS and KRAS showed dysplastic neutrophils and megakaryocytes (insert), and increased basophils and blasts. Original magnification: ×1000. B Bone marrow from a patient with AML with t(6;9) and FLT3-ITD mutation showed trilineage dysplasia and increased myeloblasts. The patient had a history of MDS-EB2 with t(6;9). Insert: dysplastic megakaryocytes. Original magnification: ×1000. C Bone marrow from a patient with AML with t(6;9) and FLT3-ITD mutation showed numerous large monoblasts. The patient developed AML progression after HSCT for MDS-EB1 with t(6;9). Original magnification: ×1000. D Bone marrow from a patient with de novo AML with t(6;9) showed numerous medium-sized myeloblasts in a background of increased basophils and dysgranulopoiesis. Original magnification: ×1000.
Immunophenotypic findings
The blasts had a non-specific myeloid immunophenotype, with consistent expression of CD13 (64/64, 100%), CD33 (64/65, 99%), CD34 (60/65, 92%), CD38 (50/50, 100%), CD117 (63/64, 98%), CD123 (21/22, 96%), HLA-DR (61/61, 100%), and MPO (36/39, 92%). Some cases expressed CD9 (5/8, 63%), CD15 (13/33, 39%), and/or monocyte-associated antigens including CD4 (9/28, 32%), CD14 (8/46, 17%), CD36 (6/13, 46%), and CD64 (39/57, 68%). Most AML cases were negative for lymphoid antigens. Cytoplasmic CD3 (n = 47) and CD5 (n = 36) were negative in all cases assessed. CD2 was positive (3/45 cases positive, 7%), CD7 (12/56, 21%), and CD19 (6/51, 12%). No AML cases in our cohort met the criteria for a mixed-phenotype acute leukemia. The blasts in most AML cases were negative for CD10 (1/39 cases positive, 3%), CD41 (3/30, 10%), and CD56 (2/44, 5%).
Cytogenetic findings
Complete karyotype results at time of initial diagnosis were available in 92 patients, including 66 with AML and 26 with MDS (Fig. 2 and Table 1). Among these patients, 67 (73%), including 45 AML and 22 MDS, had t(6;9) or its variant translocation as a sole cytogenetic abnormality, including 1 MDS with t(6;9;15)(p23;q34;q25). The remaining 25 patients (21 AML and 4 MDS) had at least one additional cytogenetic abnormality including 9 with a complex karyotype (7 AML and 2 MDS). The most common additional chromosomal aberrations included trisomy 8 (n = 9) and trisomy 13 (n = 5), all in patients with AML. In four cases of MDS with additional abnormalities, del[15q] and a complex karyotype were observed in two patients each. No significant differences were observed in the frequencies of a complex karyotype or sole t(6;9) between the MDS vs. AML groups (p > 0.99 and 0.13, respectively).
Twenty-four cases with t(6;9), including 13 AML and 11 MDS, had FISH performed to confirm the DEK-NUP214. All cases with positive DEK-NUP214 by FISH had their t(6;9) detected by karyotype. In addition, 22 cases, including 13 AML and 9 MDS, were assessed by RT-PCR using an acute leukemia translocation panel. Twenty-one patients were positive for DEK-NUP214 fusion transcripts. The only MDS patient negative for the fusion transcript was confirmed to carry DEK-NUP214 by FISH.
Of the nine patients without complete karyotype information at the time of initial presentation, two had AML with t(6;9)/DEK-NUP214 detected by FISH, and seven had t(6;9) but a complete karyotype was not available (four AML and three MDS). Two MDS patients had a normal karyotype with cryptic DEK-NUP214 fusion detected by the acute leukemia translocation panel (n = 1) or FISH (n = 1) testing.
Molecular findings by targeting sequencing
Molecular profiles (except FLT3 described separately) at initial diagnosis were available in 58 patients: 44 with AML and 14 with MDS (Supplementary Table 1).
In cases of AML with t(6;9)/DEK-NUP214, RAS genes were mutated in 12 of 33 (36%) cases assessed including 11 NRAS and 1 HRAS. Other gene mutations in AML with t(6;9)/DEK-NUP214 included TET2 (3/16, 19%), IDH2 (3/20, 15%), PTPN11 (2/13, 15%), EGFR (1/8, 13%), SETBP2 (1/8, 13%), WT1 (2/16, 13%), PRPF40B (1/9, 11%), U2AF1 (1/9, 11%), DNMT3A (1/17, 6%), IDH1 (1/18, 6%), and NPM1 (1/29, 3%). No mutations were detected in ASXL1 (n = 9), BCORL1 (n = 6), CEBPA (n = 28), JAK2 (n = 24), KIT (n = 26), KMT2A (n = 5), NOTCH1 (n = 11), PHF6 (n = 11), PTEN (n = 14), and ZRSR2 (n = 6).
In cases of MDS with t(6;9)/DEK-NUP214, RAS was mutated in five of ten (50%) patients assessed (four NRAS and one KRAS). Other genes mutated in MDS with t(6;9)/DEK-NUP214 included DNMT3A (2/7, 29%), WT1 (2/8, 25%), PTPN11 (1/4, 25%), ZRSR2 (1/4, 25%), PHF6 (1/5, 20%), TET2 (1/7, 14%), and IDH1 (1/8, 13%). No mutations were found in ASXL1 (n = 7), BCORL1 (n = 4), CEBPA (n = 4), KIT (n = 8), KMT2A (n = 2), IDH2 (n = 8), NOTCH1 (n = 5), and NPM1 (n = 8).
FLT3-ITD mutation was detected in 61 of 79 (77%) cases assessed (Table 1). The frequency of FLT3-ITD mutation in MDS was lower than that in AML (10/19, 53% vs. 51/60, 85%, p = 0.01). Twelve cases of AML and one MDS had a FLT3-ITD ratio available, and four cases including the MDS case showed multiple peaks, whereas the remaining AML cases had a median ratio of 0.042 (range, 0.012–0.782). The median interval for patients with MDS associated with FLT3-ITD to develop AML was 16.1 months (range, 0.7–27.4). The three MDS patients without FLT3-ITD developed AML after 2.3, 9.5, and 20.7 months. Kaplan–Meier analysis for time interval from MDS diagnosis to AML progression was performed in the MDS group with FLT3-ITD mutation vs. without FLT3-ITD mutation; there was no difference in AML progression between these two groups (p = 0.2). In addition, five patients were found to have FLT3 D835 mutations (three with MDS and two with AML).
Clinical outcomes
The median follow-up time was 24 months for the entire cohort (range, 1–160 months). After a median interval of 13 months (range, 1–43 months), 20 of 33 (61%) patients with MDS developed AML. Most of patients in this study received the 7 + 3 regimen during their disease course, including 63 of 72 (88%) with AML and 24 of 33 (73%) with MDS (p = 0.09). In the 24 MDS patients treated with the 7 + 3 regimen, 5 patients were treated during their MDS disease phase, 14 patients during their AML disease phase, and 5 patients did not develop AML during their follow-up period. As expected, more patients with MDS received hypomethylating agent treatment than patients with AML (16/33, 49% vs. 12/72, 17%, p = 0.0016). Sixty-nine (64%) patients received HSCT during their clinical course with very similar rates between patients with MDS (20/33, 61%) and patients with AML (49/74, 66%) (p = 0.66).
At time of last follow-up, 43 of 74 (58%) AML and 17 of 33 (52%) MDS patients died. There was no difference in OS between patients with MDS and patients with AML (median, 39 vs. 24 months, p = 0.18; Fig. 3A). When further stratified by the status of transplantation, patients who received HSCT had a longer OS than those without HSCT (median, 62 vs. 17 months, p < 0.0001; Fig. 3B). Without HSCT, patients with MDS had a better OS than patients with AML (median, 26 vs. 13 months, p = 0.0035; Fig. 3C). With HSCT, MDS and AML patients had a similar survival (median, not reached vs. 48 months, p = 0.14; Fig. 3D). In addition, to evaluate the effect of induction therapy on OS of patients with t(6;9)/DEK-NUP214, we compared patients who received 7 + 3 chemotherapy vs. those who did not. There was no statistically significant difference between these groups (p = 0.4).
A No difference in OS between patients with MDS vs. patients with AML were noted (median: 39 vs. 24 months, p = 0.18). B Patients who received HSCT had a longer OS than patients who did not (median: 62 vs. 17 months, p < 0.0001). C Without HSCT, MDS patients with t(6;9) showed a better survival than AML patients with t(6;9) (median: 26 vs. 13 months, p = 0.0035). D With HSCT, MDS patients with t(6;9) showed a similar survival than AML patients with t(6;9) (median: not reached vs. 48 months, p = 0.14).
Multivariate analyses using Cox-regression models were used to evaluate prognostic factors for OS in patients with MDS or AML with t(6;9)/DEK-NUP214. In an initial model that included 16 variables (see Statistical analysis in Materials and Methods), only initial diagnosis (MDS vs. AML) and transplantation were significant prognostic factors (p = 0.008 and p < 0.0001, respectively; Table 2). However, similar to the OS analysis, initial diagnosis of MDS vs. AML had a prognostic impact only in patients who were not treated with transplantation (p = 0.005; Table 2).
In addition, we compared OS of AML patients with t(6;9)/DEK-NUP214 in our cohort to a more general AML patients in a TCGA cohort, which utilized the European LeukemiaNet standardized system for cytogenetic subtypes of AML [27, 28]. The analyses showed that the OS of AML patients with t(6;9)/DEK-NUP214 was similar to that of AML patients with intermediate cytogenetic risk (median, 24 vs. 27 months, p = 0.7), slightly better than that with unfavorable cytogenetic risk (median, 24 vs. 12 months, p = 0.04), and significantly worse than that with favorable cytogenetic risk (median, 24 vs. not reached, p = 0.02) in the TCGA database.
Discussion
Myeloid neoplasms with t(6;9)/DEK-NUP214 are rare. In over two decades, only 50 cases of AML and 16 cases of MDS were identified from at MDACC where over 18,000 BM specimens are processed annually. Although AML with t(6;9)/DEK-NUP214 is described as a distinct entity in the WHO classification, many aspects of this disease remain unknown, especially prognostic factors. MDS with t(6;9)/DEK-NUP214 has been studied even less. One cohort was reported in which the clinicopathologic features between t(6;9)/DEK-NUP214-positive MDS and AML were compared; the authors suggested that myeloid neoplasms with t(6;9)/DEK-NUP214 may be best categorized as AML, irrespective of the blast percentage [9]. However, this study only included eight MDS patients and focused on a pediatric population. To the best of our knowledge, our study represents the largest cohort to characterize and compare the clinicopathologic features between MDS and AML associated with t(6;9)/DEK-NUP214, including cytogenetic and molecular findings as well as clinical outcomes.
In our study, MDS patients tend to be older than AML patients. No gender predilection was present in our cohort. The laboratory findings showed that MDS patients had a lower white blood cell count and a higher platelet count than AML patients; hemoglobin levels were not different. These data indicate that MDS patients had a lesser tumor burden than AML patients, likely attributable to their differences in blast count. Previous studies [1, 7] showed that AML with t(6;9)/DEK-NUP214 is morphologically and immunophenotypically similar to AML with maturation or acute myelomonocytic leukemia, which was confirmed in our study. In our cohort, about 60% of AML cases showed evidence of multilineage dysplasia, which may be a morphologic hint for the presence of t(6;9)/DEK-NUP214, although the frequency of multilineage dysplasia was lower when compared to MDS cases. Although some studies described basophilia as a pathologic feature in AML with t(6;9)/DEK-NUP214, basophilia was not present in neither the PB nor BM in most patients in this cohort, consistent with more recent publications [8,9,10].
The cytogenetic data in this study show that about three quarters of patients with MDS or AML had t(6;9) as an isolated abnormality; only 10% had a complex karyotype. These results are in line with earlier studies and suggest that t(6;9)/DEK-NUP214 is a main driver in pathogenesis. Similar to an earlier study [12], trisomy 13 and trisomy 8 were the common additional abnormalities observed in cases without a complex karyotype in this cohort. Similar to previous studies [9, 12], the results in this study show that patients with t(6;9)/DEK-NUP214 frequently harbor a concomitant FLT3-ITD, although the rate was lower in MDS than AML. Multivariate analyses showed that FLT3-ITD is not a significant adverse prognostic factor for OS, concurring with some other studies [9, 10, 12]. RAS mutation, especially NRAS, was another frequent abnormality in both MDS and AML patients with t(6;9)/DEK-NUP214, as was reported by Visconte et al. [10]. Due to the inevitable limitation of a retrospective study, molecular profiles were not complete in our cohort. Further studies are necessary to elucidate the spectrum of molecular abnormalities in patients with MDS or AML associated with t(6;9)/DEK-NUP214.
Over half of the patients in this cohort died with a median follow-up of 24 months, despite being treated with aggressive chemotherapy regimens and HSCT. There was no difference in OS between MDS and AML patients in the entire cohort, but the survival curves did show a trend toward favorable survival in MDS patients at the earlier stage of diseases (Fig. 3A). The multivariate analysis on all patients with t(6;9)/DEK-NUP214 confirmed the prognostic significance of the initial diagnosis of MDS vs. AML after adjusting 15 potential confounding factors (Table 2). These results argue against the proposal that MDS associated with t(6;9)/DEK-NUP214 should be classified as AML irrespective of blast percentage. Our data confirm previous studies [12, 29,30,31] showing that HSCT significantly improve the survival of patients with myeloid neoplasms with t(6;9)/DEK-NUP214: 62 months of median survival in patients who received HSCT vs. 17 months in patients who did not (p < 0.0001). In contrast, induction therapy appears to have no effect on the survival (p = 0.4). Given the significant prognostic effect of HSCT in these patients, we performed further analyses by stratification according to transplantation status. Multivariate analyses showed that MDS patients with t(6;9)/DEK-NUP214 have a better survival than AML patients with t(6;9)/DEK-NUP214 among non-transplanted patients. In transplanted patients, OS was not significantly different, indicating that HSCT greatly improves clinical outcomes and may overcome the adverse effect of the initial diagnosis of AML on the survival.
In earlier studies, patients with AML associated with t(6;9)/DEK-NUP214 were thought to have a poor prognosis. In a cohort including 69 AML patients with t(6;9) from 1987 to 2002 [1], the median survival was 14.4 months, worse than the median survival time of 24 months in our cohort. However, HSCT was performed in only 31% of their patients vs. 66% in our cohort. Another cohort of 178 AML patients with t(6;9) from 1989 to 2016 had a median OS of 27 months with a transplantation rate of 62%, which are comparable to our findings [12]. As our data has shown, transplantation has a significant prognostic effect on these patients, and may explain the differences in survival time between these studies. In addition, our analysis comparing our cohort with the TCGA AML database shows that patients with AML associated with t(6;9)/DEK-NUP214 that receive aggressive treatment strategies have a prognosis similar to AML patients in the intermediate-risk cytogenetic group. These data suggest that the poor prognosis of this disease may be overcome with modern intensive therapy.
In our opinion, MDS with t(6;9)/DEK-NUP214 does share some clinicopathologic characteristics with AML with t(6;9)/DEK-NUP214, including a comparable hemoglobin level, frequent multilineage dysplasia, FLT3-ITD, RAS mutation, and a high frequency of t(6;9)/DEK-NUP214 as an isolated abnormality. However, MDS and AML patients also differ in some aspects, especially the differences on OS. Therefore, we suggest that MDS with t(6;9)/DEK-NUP214 should not be categorized with AML with t(6;9)/DEK-NUP214. However, given that over half of MDS patients with t(6;9)/DEK-NUP214 developed AML, and that transplantation can greatly improve their clinical outcomes, early and aggressive treatment strategies such as HSCT may be beneficial for MDS patients associated with t(6;9)/DEK-NUP214.
References
Slovak ML, Gundacker H, Bloomfield CD, Dewald G, Appelbaum FR, Larson RA, et al. A retrospective study of 69 patients with t(6;9)(p23;q34) AML emphasizes the need for a prospective, multicenter initiative for rare ‘poor prognosis’ myeloid malignancies. Leukemia. 2006;20:1295–7.
Arber DA, Brunning RD, Le Beau MM, Falini B, Vardiman JW, Porwit A, et al. Acute myeloid leukaemia with t(6;9)(p23;q34.1); DEK-NUP214. In: Swerdlow S, Campo E, Harris N, Jaffe E, Pileri S, Stein H, et al., editors. WHO classification of tumours of haematopoietic and lymphoid tissues. Revised 4th ed. Lyon: IARC; 2017. p. 137–8.
Arber DA, Brunning RD, Le Beau MM, Falini B, Vardiman JW, Porwit A, et al. Acute myeloid laeukemia with t(6;9)(p23;q34); DEK-NUP214. In: Swerdlow S, Campo E, Harris N, Jaffe E, Pileri S, Stein H, et al., editors. WHO classification of tumours of haematopietic and lymphoid tissues. 4th ed. Lyon: IARC; 2008. p. 115–6.
Rowley JD, Potter D. Chromosomal banding patterns in acute nonlymphocytic leukemia. Blood. 1976;47:705–21.
Boer J, Bonten-Surtel J, Grosveld G. Overexpression of the nucleoporin CAN/NUP214 induces growth arrest, nucleocytoplasmic transport defects, and apoptosis. Mol Cell Biol. 1998;18:1236–47.
Ageberg M, Drott K, Olofsson T, Gullberg U, Lindmark A. Identification of a novel and myeloid specific role of the leukemia-associated fusion protein DEK-NUP214 leading to increased protein synthesis. Genes Chromosomes Cancer. 2008;47:276–87.
Oyarzo MP, Lin P, Glassman A, Bueso-Ramos CE, Luthra R, Medeiros LJ. Acute myeloid leukemia with t(6;9)(p23;q34) is associated with dysplasia and a high frequency of flt3 gene mutations. Am J Clin Pathol. 2004;122:348–58.
Gupta M, Ashok Kumar J, Sitaram U, Neeraj S, Nancy A, Balasubramanian P, et al. The t(6;9)(p22;q34) in myeloid neoplasms: a retrospective study of 16 cases. Cancer Genet Cytogenet. 2010;203:297–302.
Sandahl JD, Coenen EA, Forestier E, Harbott J, Johansson B, Kerndrup G, et al. t(6;9)(p22;q34)/DEK-NUP214-rearranged pediatric myeloid leukemia: an international study of 62 patients. Haematologica. 2014;99:865–72.
Visconte V, Shetty S, Przychodzen B, Hirsch C, Bodo J, Maciejewski JP, et al. Clinicopathologic and molecular characterization of myeloid neoplasms with isolated t(6;9)(p23;q34). Int J Lab Hematol. 2017;39:409–17.
Tarlock K, Alonzo TA, Moraleda PP, Gerbing RB, Raimondi SC, Hirsch BA, et al. Acute myeloid leukaemia (AML) with t(6;9)(p23;q34) is associated with poor outcome in childhood AML regardless of FLT3-ITD status: a report from the Children’s Oncology Group. Br J Haematol. 2014;166:254–9.
Kayser S, Hills RK, Luskin MR, Brunner AM, Terre C, Westermann J, et al. Allogeneic hematopoietic cell transplantation improves outcome of adults with t(6;9) acute myeloid leukemia: results from an international collaborative study. Haematologica. 2020;105:161–9.
Chi Y, Lindgren V, Quigley S, Gaitonde S. Acute myelogenous leukemia with t(6;9)(p23;q34) and marrow basophilia: an overview. Arch Pathol Lab Med. 2008;132:1835–7.
Fan YS, Raza A, Schumer J, Sait SN, Block AW, Snyderman M, et al. Translocation t(6;9)(p22.3;q34) in myelodysplastic syndrome-refractory anemia with excess blasts. Cancer Genet Cytogenet. 1987;29:135–8.
Cuneo A, Kerim S, Vandenberghe E, Van Orshoven A, Rodhain J, Bosly A, et al. Translocation t(6;9) occurring in acute myelofibrosis, myelodysplastic syndrome, and acute nonlymphocytic leukemia suggests multipotent stem cell involvement. Cancer Genet Cytogenet. 1989;42:209–19.
Soekarman D, von Lindern M, Daenen S, de Jong B, Fonatsch C, Heinze B, et al. The translocation (6;9) (p23;q34) shows consistent rearrangement of two genes and defines a myeloproliferative disorder with specific clinical features. Blood. 1992;79:2990–7.
Shapira MY, Hirshberg B, Amir G, Rund D. 6;9 translocation in myelodysplastic syndrome. Cancer Genet Cytogenet. 1999;112:57–9.
Ferro MT, Resino M, Cabello P, Lopez-Yarto A, Mazariego YV, Garcia-Sagredo JM, et al. t(6;9)(p22.3;q34) in a patient with refractory anemia with excess of blasts in transformation. Cancer Genet Cytogenet. 1993;69:74–5.
Gold EJ, Conjalka M, Pelus LM, Jhanwar SC, Broxmeyer H, Middleton AB, et al. Marrow cytogenetic and cell-culture analyses of the myelodysplastic syndromes: insights to pathophysiology and prognosis. J Clin Oncol. 1983;1:627–34.
Yunis JJ. Recurrent chromosomal defects are found in most patients with acute nonlymphocytic leukemia. Cancer Genet Cytogenet. 1984;11:125–37.
Quesada AE, Routbort MJ, DiNardo CD, Bueso-Ramos CE, Kanagal-Shamanna R, Khoury JD, et al. DDX41 mutations in myeloid neoplasms are associated with male gender, TP53 mutations and high-risk disease. Am J Hematol. 2019;94:757–66.
Patel KP, Ruiz-Cordero R, Chen W, Routbort MJ, Floyd K, Rodriguez S, et al. Ultra-rapid reporting of GENomic targets (URGENTseq): clinical next-generation sequencing results within 48 h of sample collection. J Mol Diagn. 2019;21:89–98.
Wang SA, Tam W, Tsai AG, Arber DA, Hasserjian RP, Geyer JT, et al. Targeted next-generation sequencing identifies a subset of idiopathic hypereosinophilic syndrome with features similar to chronic eosinophilic leukemia, not otherwise specified. Mod Pathol. 2016;29:854–64.
Warren M, Luthra R, Yin CC, Ravandi F, Cortes JE, Kantarjian HM, et al. Clinical impact of change of FLT3 mutation status in acute myeloid leukemia patients. Mod Pathol. 2012;25:1405–12.
Patel KP, Barkoh BA, Chen Z, Ma D, Reddy N, Medeiros LJ, et al. Diagnostic testing for IDH1 and IDH2 variants in acute myeloid leukemia an algorithmic approach using high-resolution melting curve analysis. J Mol Diagn. 2011;13:678–86.
Luthra R, Patel KP, Reddy NG, Haghshenas V, Routbort MJ, Harmon MA, et al. Next-generation sequencing-based multigene mutational screening for acute myeloid leukemia using MiSeq: applicability for diagnostics and disease monitoring. Haematologica. 2014;99:465–73.
Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74.
Mrozek K, Marcucci G, Nicolet D, Maharry KS, Becker H, Whitman SP, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012;30:4515–23.
Ishiyama K, Takami A, Kanda Y, Nakao S, Hidaka M, Maeda T, et al. Allogeneic hematopoietic stem cell transplantation for acute myeloid leukemia with t(6;9)(p23;q34) dramatically improves the patient prognosis: a matched-pair analysis. Leukemia. 2012;26:461–4.
Ishiyama K, Takami A, Kanda Y, Nakao S, Hidaka M, Maeda T, et al. Prognostic factors for acute myeloid leukemia patients with t(6;9)(p23;q34) who underwent an allogeneic hematopoietic stem cell transplant. Leukemia. 2012;26:1416–9.
Diaz-Beya M, Labopin M, Maertens J, Alijurf M, Passweg J, Dietrich B, et al. Allogeneic stem cell transplantation in AML with t(6;9)(p23;q34);DEK-NUP214 shows a favourable outcome when performed in first complete remission. Br J Haematol. 2020;189:920–5.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Fang, H., Yabe, M., Zhang, X. et al. Myelodysplastic syndrome with t(6;9)(p22;q34.1)/DEK-NUP214 better classified as acute myeloid leukemia? A multicenter study of 107 cases. Mod Pathol 34, 1143–1152 (2021). https://doi.org/10.1038/s41379-021-00741-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-021-00741-w
This article is cited by
-
The International Consensus Classification of myelodysplastic syndromes and related entities
Virchows Archiv (2023)