- Molecular Cell Biology, University of Groningen, Groningen, Netherlands
We have evaluated the current knowledge on peroxisome proliferation in yeast. In wild-type cells, peroxisomes multiply predominantly by fission at conditions that require peroxisome function(s) for growth. In cells that lack peroxisomes, for instance in pex3 and pex19 mutants or in mutants that display inheritance defects, peroxisomes may form de novo. We propose a novel machinery for the de novo formation of peroxisomes in pex3 cells, in which new peroxisomes do not arise from the endoplasmic reticulum. This machinery is based on the recent observation that membrane vesicles are present in pex3 cells that display peroxisomal characteristics in that they contain specific peroxisomal membrane and matrix proteins. These structures are the source for newly formed peroxisomes upon reintroduction of Pex3. Furthermore, we critically evaluate the principles of sorting of other peroxisomal membrane proteins to their target organelle and the function of the endoplasmic reticulum therein.
Introduction
Peroxisomes are highly versatile organelles that readily adapt their numbers and physiological function in relation to metabolic needs. This functional flexibility requires a careful regulation of controlling organelle number and size. The factors controlling organelle size are still an enigma.
In yeast, low numbers of peroxisomes are normally present in cells grown at glucose excess conditions. However, when the cells are placed in media supplemented with carbon sources that require peroxisomal enzymes for growth (i.e., fatty acids, methanol, purines, and D-amino acids), organelle proliferation rapidly starts (Veenhuis et al., 1978). The mode of yeast peroxisome multiplication is still controversial. The current models range from the suggestion that in normal wild-type (WT) cells peroxisome multiplication exclusively results from fission to the view that all organelles form de novo from the endoplasmic reticulum (ER). Also combinations of these two modes have been suggested.
This contribution presents a critical overview of recent data on peroxisome multiplication in yeast and proposes possible novel directions aimed at resolving the molecular mechanisms of peroxisome formation.
Peroxisome Development
The origin of peroxisomes is still controversial. Following their discovery, the organelles were considered to bud from the ER based on the observations that peroxisomes—and in particular young developing ones—were invariably seen in close contact with the ER. Since direct contacts between the organelles were not observed, the ER theory was replaced by a model of development by growth and fission of pre-existing ones (Lazarow and Fujiki, 1985). The first morphological data that suggested growth and fission came from kinetic studies using the yeast Hansenula polymorpha, shifted from glucose to methanol, conditions that require peroxisome enzymes for growth (Veenhuis et al., 1978). During growth on glucose, H. polymorpha cells contain a single peroxisome. In the first 6–8 h after the shift alcohol oxidase and catalase, key peroxisomal enzymes of methanol metabolism, are synthesized and incorporate in the original organelles present in the glucose inoculum cells. After maturation, the organelle formed an extension that subsequently budded off and in turn grew (Figure 1). This way the cells formed 5–7 organelles of approximately equal size (Figure 1) within a period of 24 h of growth. These morphological data were subsequently reinforced by biochemical data which indicated that peroxisomal matrix proteins were synthesized on free ribosomes in the cytosol (Goldman and Blobel, 1978; Fujiki et al., 1985) and post-translationally incorporated in the organelle by a unique protein translocation machinery (Lazarow and Fujiki, 1985).
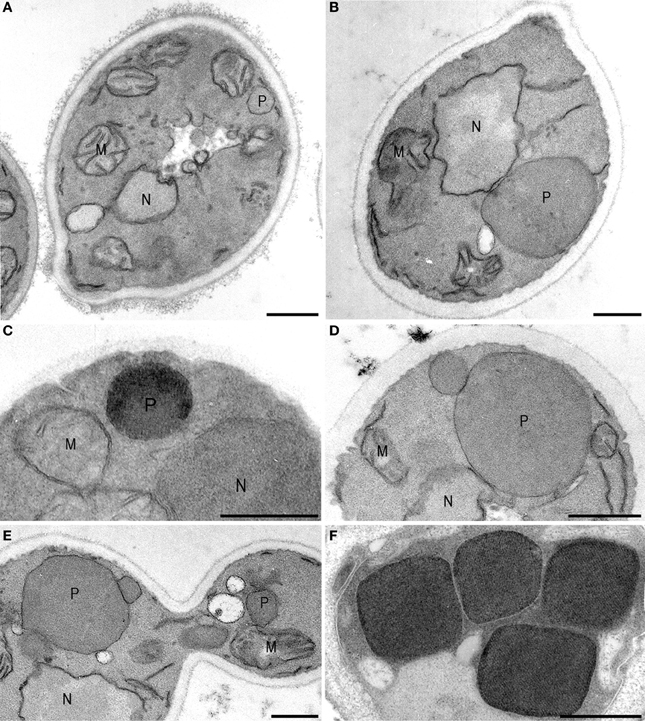
Figure 1. Peroxisome multiplication imaged. Hansenula polymorpha cells are shifted from glucose- to methanol media, conditions that require peroxisome functions for growth. On glucose, characteristically a single peroxisome is present per cell (A) that—upon a shift to methanol containing media—incorporates alcohol oxidase (C), catalase and dihydroxyacetone synthase proteins, which are essential for growth on methanol. As a result the organelle increase in size (B) and, after maturation, forms a new organelle by fission (D) that subsequently will grow. A similar a-symmetric fission machinery is responsible for the administration of small organelles to the yeast bud (E). When the culture has reached the late exponential phase of growth, typically 4–6 organelles are present of comparable shape. The cuboid shape is due to the presence of large luminal alcohol oxidase crystals. M, mitochondria; N, nucleus; P, peroxisome. Cells are fixed with KMnO4, except (C,F), which are glutaraldehyde fixed. In these cells alcohol oxidase activity is demonstrated using CeCl3.
The first genetic support for the growth and fission model came from studies in the Tabak group, who identified the Saccharomyces cerevisiae dynamin-like protein (DLPs) Vps1 as a component involved in peroxisome fission together with actin and the class V myosin motor Myo2 being required for transport of newly separated organelles to the developing bud (Hoepfner et al., 2001). Studies in other yeast species, plant and mammals resolved another DLPs (designated Drp1, Dnm1 or DRP3 respectively) that is involved in peroxisome fission (Koch et al., 2003; Kuravi et al., 2006; Zhang and Hu, 2009). Interestingly, these proteins also play a role in mitochondrial fission. Subsequent studies identified additional components, namely the tail anchored protein Fis1 that, together with Mdv1 (only in yeast) and, unique for S. cerevisiae, Caf1, are required to bind Dnm1—but not Vps1—to the target membrane (Motley et al., 2008; Nagotu et al., 2008a). In addition, Mff1 and GDAP1 have been identified to control both mitochondria and peroxisome fission in mammalian cells (Gandre-Babbe and van der Bliek, 2008; Huber et al., 2013). With this, consensus had been reached in the field for the autonomous nature of peroxisomes for many years.
This view changed again when the first data came available on the reintroduction of peroxisomes in cells lacking the organelles due to a mutation in a gene essential for peroxisome membrane biogenesis. Bulk of these studies were conducted with pex3 or pex19 cells, in which the organelles reappeared after reintroduction of the corresponding deleted gene, by a process often referred to as “de novo peroxisome formation” (Hoepfner et al., 2005; Kragt et al., 2005; Tam et al., 2005; Haan et al., 2006). It is commonly accepted that peroxisomes, which form de novo, are not created from scratch, but originate from another membrane in line with the proposition “Omnis membrana e membrana” (Günther Blobel, Nobel Prize 1999). Most of the available experimental data point to the ER as a template for this pathway.
With this, the question raised whether and in how far this process contributes to the total peroxisome population in WT cells. Indications for this came from studies in which VPS1 and/or DNM1 were deleted. In all yeast species studied this resulted in the reduction of peroxisome numbers to generally only one organelle per cell. Under these conditions the de novo peroxisome formation machinery is normally active since Dnm1 and Vps1 are not involved in this process (Motley et al., 2008; Nagotu et al., 2008b). Very recently, it was shown that a peroxisome-deficient phenotype was obtained in mutant yeast cells in which both de novo synthesis and fission are blocked (in H. polymorpha pex11 pex25 cells), but not when only one of these processes was blocked in pex11 or pex25 cells; (Saraya et al., 2011). This reinforces that in yeast the cellular peroxisome population can be maintained predominantly by fission (in pex25 cells).
Taken all data together, fission appears to be the dominant mode of organelle maintenance in yeast although it cannot be excluded that few organelles are formed de novo too in WT cells. Evidence for the latter is however not observed. Possibly, the yeast model is not universal as data have been presented suggesting that in mammals de novo synthesis prevails in organelle formation (Kim et al., 2006). However, other studies indicate that mammalian peroxisomes also predominantly form by fission (Huybrechts et al., 2009; Delille et al., 2010).
Clearly, organelle multiplication in substrate induced yeast cells serves different functions: in WT cells new organelles will be formed that will stay in the mother cell and mature to support growth on the carbon source that is supplemented for growth (i.e., oleate or methanol). On the other hand, organelles multiply dependent of the cell cycle to administrate new organelles to the daughter cell (Figure 2). While mother organelles are hooked up in the mother cell via the function of Inp1, these newly formed organelles bind Inp2, which is required for binding of the organelle to Myo2 and subsequent transport to the developing bud (Fagarasanu et al., 2005, 2006; Knoblach et al., 2013). This suggests that upon peroxisome fission two types of organelles may form that biochemically differ. Possibly, this is related to the function of Pex19. This suggestion is based on the important observation that Inp2 is not the sole determinant in Myo2 binding in that Inp2 interacts with both Myo2 and Pex19 to serve the function in organelle transport to the bud (Otzen et al., 2012). Therefore, it may well be that the availability of Pex19 at the membrane (and thus Inp2 binding) is the key determinant that prescribes which organelle is donated to the bud and which one will stay in the mother cell to serve a function in optimal cell metabolism.
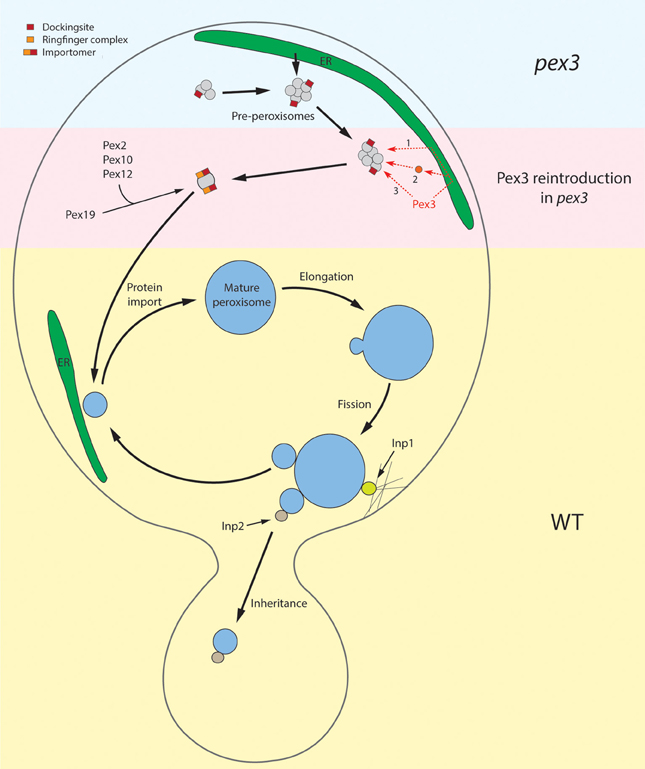
Figure 2. Schematic overview of peroxisome re-introduction in pex3 and peroxisome proliferation in WT cells. The upper part of the cell represents the situation in pex3 cells. These cells contain vesicular structures that harbor Pex13 and Pex14, proteins of the receptor docking site. How these proteins are sorted to these membranes is unknown. These structures may derive by proliferation of pre-existing ones or be formed from the ER. The pre-peroxisomes are the target for re-introduced Pex3, which may reach these structures via the ER (1), upon incorporation in vesicles that derive from the ER and subsequently fuse with pre-peroxisomes (2) or sort directly to them (3). With the presence of Pex3 at the pre-peroxisome, other PMPs can subsequently be taken up via the Pex3/Pex19 machinery. These include the ring finger proteins Pex2, Pex10, and Pex12, leading to the formation of a functional importomer. This allows uptake of matrix proteins resulting in organelle maturation and subsequent multiplication by fission as depicted in the lower half of the picture (WT situation). During growth the organelle is invariably closely associated with strands of ER. During cell fission Inp1 is essential to dock the mother organelle to the cortex whereas Inp2 determines the delivery of newly formed organelles to the bud.
Obviously, peroxisome fission and partition have to be carefully controlled to sustain optimal cell health. Indeed, a mutation in human DLP1 results in a lethal phenotype (Waterham et al., 2007). The fission process can be divided into three steps, namely the initial organelle elongation step, growth and constriction of the elongated organelle followed by the actual fission process. Various proteins have been suggested to be involved in peroxisome fission, including members of the Pex11 and the Pex23 protein families (Kiel et al., 2006). In yeast the molecular function of most of these proteins is unknown except for Pex11 and Pex25 (Saraya et al., 2011). Opalinski and colleagues demonstrated that Pex11 is specifically involved in the initial membrane elongation process, a function that is mediated by an amphipathic α-helix located in the N-terminus of the protein and that is conserved between species (Opalinski et al., 2011). The principles of constriction are unknown but may, similar to mitochondria, be related to the function of the ER in conjunction with actin filaments. Scission is mediated by Dlp's, i.e., Vps1 and Dnm1 in baker's yeast (Hoepfner et al., 2001; Kuravi et al., 2006) and Dnm1 in H. polymorpha (Nagotu et al., 2008b).
Surprisingly, peroxisome fission is associated with a major rearrangement of various peroxisomal membrane proteins (PMPs) belonging to the importomer that are specifically donated to the developing small organelle (Cepinska et al., 2011). This process appeared to be dependent on the function of Pex11. In contrast to the bulk administration of PMPs to the small peroxisome, fluorescence microscopy analysis revealed that generally very low levels of matrix components, often below the limit of detection, are included in these structures. Apparently, during yeast fission the developing bud is administrated with new organelles that are optimally equipped for their function in matrix protein import (thereby determining their future function!) rather than having an immediate function in cell metabolism (Figure 2).
Obviously, peroxisome development requires functional interactions with other organelles, such as the ER, where most peroxisomal phospholipids are synthesized (Raychaudhuri and Prinz, 2008) as well as with mitochondria, which synthesize cardiolipin, a lipid present in the peroxisomal membrane (Wriessnegger et al., 2007), and heme (for catalase synthesis). So far, very little is known on possible physical interactions of peroxisomes and other organelles, which may contribute to various aspects of peroxisome proliferation.
De novo Peroxisome Formation
Peroxisome Reintroduction in pex3 Yeast Cells
The most extensively used experimental system to study de novo peroxisome formation in yeast is the reintroduction of peroxisomes in pex3 strains in which a PEX3-GFP hybrid gene is placed under control of an inducible promoter (Hoepfner et al., 2005; Kragt et al., 2005; Tam et al., 2005; Haan et al., 2006). Fluorescence microscopy (FM) analysis revealed that upon induction newly synthesized Pex3-GFP sorts to the ER, concentrates in foci followed by the formation of a pre-peroxisomal structure, which pinches off and develops into a nascent peroxisome. Alternatively two (S. cerevisiae; van der Zand et al., 2012) or multiple (Y. lipolytica; Titorenko et al., 2000) types of vesicles have been proposed to pinch off from the ER, which subsequently fuse to form a nascent peroxisome.
Invariably, these models predict that Pex3 initially sorts to the ER. This is strengthened by the finding that the extreme N-terminus of Pex3 proteins has characteristics in common with signal-anchor-like sequences, typical for authentic ER membrane proteins (Thoms et al., 2012; Fakieh et al., 2013). Another argument for ER sorting of Pex3 is the observation that peroxisome reintroduction in S. cerevisiae pex3 cells is affected in a temperature sensitive sec61 mutant strain or upon depletion of Sec61 (Thoms et al., 2012). However, opposite data were described by the Gould group (South et al., 2001) using a cold-sensitive S. cerevisiae sec61 mutant.
Also, experiments reported by Kragt and colleagues suggest initial sorting of Pex3 to the ER (Kragt et al., 2005). In this study a Pex3 variant was used containing an artificial ER signal peptide and cleavage site, which functionally complemented S. cerevisiae pex3 cells, whereas a similar construct in which cleavage of the signal peptide was blocked by a mutation did not. This result was interpreted by an inability of the uncleaved variant to exit the ER during de novo peroxisome formation.
However, these studies and other studies were conducted at conditions in which the N-terminus of Pex3 was modified or in which PEX3 was not under control of its endogenous promoter, which could lead to artificial mislocalization.
Analysis of mRNA transcript levels demonstrated that the expression of genes encoding peroxins/PMPs hardly increased at peroxisome induction conditions (Kal et al., 1999). Hence, overproduction is readily achieved which may lead to mislocalization. This may also be true for Pex3, when produced under control of strong inducible promoters, even when the ultimate Pex3 levels do not exceed WT levels. This is related to the fact that the initial kinetics of the PGAL driven Pex3 synthesis strongly exceeds those normally occurring in WT cells via the endogenous promoter.
Indeed, also data have been presented suggesting that the ER is not the target membrane for authentic Pex3. For instance, studies by Matsuzaki and Fujiki, who analyzed in vitro insertion of Pex3 in different cellular fractions of Chinese hamster ovary cells, revealed that Pex3 only inserted in peroxisomal membranes, but not in microsomes or mitochondria (Matsuzaki and Fujiki, 2008). Similarly, in vitro experiments indicated that PMP24 was directly inserted in rat liver peroxisomes in a Pex3 and Pex19 dependent manner (Pinto et al., 2006).
Although the data reported by Kragt and colleagues, strongly suggest that Pex3 traffics via the ER (Kragt et al., 2005), a similar approach in the Erdmann group using a Pex3 variant containing artificial sorting information for the mitochondrial outer membrane protein, also resulted in functional complementation of pex3 cells (Rucktaschel et al., 2010). This at least suggests that Pex3 not necessarily needs to sort via the ER for de novo peroxisome formation. Moreover, for both experiments it cannot be excluded that a minor portion of the modified Pex3 escapes from sorting to mitochondria or ER.
Moreover, two recent studies revealed that newly synthesized Pex3 targets to all pre-existing peroxisomes in WT yeast cells (Fakieh et al., 2013; Menendez-Benito et al., 2013). If under these conditions Pex3 would traffic via the ER, a vesicular transport pathway from the ER to pre-existing peroxisomes should exist. Indeed data have been presented supporting this possibility (Motley and Hettema, 2007). On the other hand, to the best of our knowledge Pex3-GFP has never been localized to the ER in WT cells.
As is clear from the above, both data indicating that Pex3 directly sorts to peroxisomes or traffics via the ER to these organelles have been presented. A model explaining these seemingly contradicting observations could be that both at the ER and peroxisomes a Pex3 insertion machinery exists. Assuming that the ER insertion machinery has a lower affinity for newly synthesized Pex3 relative to the peroxisomal one, Pex3 will predominantly sort to the high affinity site at peroxisomes in WT cells. However, in cells lacking peroxisomes or at Pex3 overproduction conditions Pex3 will then (also) be sorted to the ER.
In vitro protein insertion studies as well as in vivo experiments tracking (single) Pex3 molecules on their way to peroxisomes in WT cells (e.g., in pulse chase experiments and/or superresolution fluorescence microscopy), will help to resolve this urgent question.
Pex3 Dependent Peroxisome Reintroduction: An Alternative Pathway
We recently re-investigated the location of Pex14 in H. polymorpha pex3 cells using high resolution immune electron microscopy and observed that the Pex14-GFP spots in fact represent vesicular structures located at the vicinity of mitochondria or ER, but never were found connected to these organelles. Comparable observations were made using H. polymorpha pex19 cells (Knoops et al., 2014). Using deconvolution microscopy, such structures previously were also observed in P. pastoris pex3 cells.
These recent studies revealed that the Pex14-containing structures in H. polymorpha pex3 cells contained, besides Pex14, also Pex8 and Pex13 and hence may contain a functional receptor docking site. However, none of the other PMPs tested (Pex10, Pmp47, Pex11) was observed at these structures, so a functional importomer was not formed. Hence, similar to matrix protein import mutants, pex3 cells apparently contain peroxisome ghosts that do not harbor all typical marker PMPs, but only a subset. This may add to the explanation why they were overlooked in earlier studies. The origin of these structures is currently unknown. We speculate that they could proliferate from pre-existing structures, like assumed for other peroxisomal ghosts. Alternatively, they may form from the ER (Figure 2). If so, however, their formation is independent of Pex3.
Interestingly, the Pex14-containing structures, but not the ER, were the target for reintroduced Pex3-GFP after which these structures developed into normal functional peroxisomes. Moreover, we also observed that Pex25 and Pex19, two other peroxins proposed to be involved in the de novo peroxisome formation, are not involved in the formation of the vesicles in pex3 cells. This new insight into peroxisome formation in pex3 cells fundamentally differs from the generally accepted models and may stimulate further studies on the principles of de novo peroxisome formation.
Peroxisome Reintroduction in Inheritance Mutants
Cells of mutants defective in peroxisome segregation (inp1 or inp2 deletion strains) temporarily lack peroxisomal structures detectable by FM (Fagarasanu et al., 2005, 2006). In these cells peroxisomes reappear shortly after budding is completed, presumably by de novo peroxisome formation (Motley and Hettema, 2007). So far, the reappearance of peroxisomes in inp1 or inp2 cells has only been studied using matrix marker proteins, but not PMP markers. Hence, it cannot be excluded yet that in these cells also small peroxisomal remnants occur, like in pex3 cells. It is important to solve this issue, using PMP marker proteins also including Pex3. If ghosts are fully absent, these mutants would be the preferred model systems for analyzing de novo peroxisome formation.
Interestingly, upon deletion of PEX25 in S. cerevisiae inp2 cells, the cells become peroxisome deficient, indicating that the de novo peroxisome formation process is blocked in these cells (Huber et al., 2012). Pex25 is also required for de novo formation in yeast pex3 cells upon reintroduction of the PEX3 gene (Saraya et al., 2011; Huber et al., 2012). These data indicate that both de novo peroxisome formation processes depend on Pex25 and probably represent the same process.
The Disputed PMP Sorting Machinery
As for the role of the ER in de novo peroxisome formation, also no consensus is reached on the involvement of the ER in trafficking of PMPs other than Pex3 to peroxisomes in WT cells. Current models range from PMP trafficking via the ER to direct post-translational insertion in peroxisomal membranes (Schliebs and Kunau, 2004; Menendez-Benito et al., 2013; Yagita et al., 2013). In the first model Pex3 and Pex19 play a role in the exit of PMPs from the ER. In the second one Pex19 serves as a soluble receptor/chaperone that binds newly synthesized PMPs and is recruited to the peroxisomal membrane by Pex3 (Fang et al., 2004), followed by the insertion of PMPs by a yet unknown mechanism. According to this model PMPs are predicted to be cytosolic or mistargeted to other cellular membranes in the absence of Pex3 or Pex19. The fact that many PMPs physically interact with Pex19 strongly supports the second model. Also, the observations that the levels of many PMPs strongly drop in pex3 cells, and often are below the limit of detection, are in favor of this model. However, also many data in support of the first model have been presented.
van der Zand and colleagues determined the localization of 16 PMPs using fluorescence microscopy upon pulsed induction using PGAL which suggested that these proteins initially sorted to the ER (van der Zand et al., 2010). As indicated above, these experiments should be interpreted with care because of the strong, initial temporal overexpression due to using PGAL. Moreover, also in these studies the limitations of the relatively low resolution of fluorescence microscopy can easily result in misinterpretation of the data. The same authors also analyzed the localization of PMPs in pex3 cells and concluded that all accumulated at the ER. Careful re-inspection of the published images suggests that Pex8, Pex13 and Pex14 indeed were present in foci, whereas the other PMPs tested (Pex2, Pex6, Pex11, Pex15) showed a very low, dispersed localization. We recently observed that also S. cerevisiae pex3 cells harbor Pex14-containing peroxisomal ghosts, like in H. polymorpha pex3 cells (unpublished results). Hence, most likely also in S. cerevisiae Pex8, Pex13 and Pex14 are present at peroxisomal membrane structures, whereas the other PMPs are instable and located to the cytosol.
In line with initial ER sorting would be a role of the Sec complex in PMP routing. Indeed, upon in vivo depletion of Sec components, a portion of certain PMPs became soluble (van der Zand et al., 2010). Also, data have been presented showing that peroxisomal tail anchor proteins depend on the function of the Get complex. For instance, Schuldiner and colleagues showed that the tail anchored protein Pex15 mislocalized to mitochondria when a component of the GET complex was depleted. Moreover, a physical interaction between Pex15 and Get complex components has been demonstrated (Schuldiner et al., 2008). Conversely, however, the insertion of the mammalian homolog of Pex15, Pex26, depends of Pex19 (Halbach et al., 2006; Matsuzono and Fujiki, 2006) and is independent of TRC40, the mammalian homolog of Get2 (Yagita et al., 2013). Also, insertion of the tail anchored protein Fis1 in peroxisomal membranes was shown to depend on Pex19 (Delille and Schrader, 2008).
As argued before for pex3 cells, in fact both pathways may exist simultaneously. In this view the final location of the PMP is determined by the affinity of it targeting information for either the ER or normal peroxisomes. An alternative may be that—at least in part—different pathways exist, depending on marker protein and model organism used.
For instance, our recent findings clearly show that the localization of Pex13 and Pex14 into peroxisomal membranes does not require Pex3. However, in the same cells other PMPs require the Pex3/Pex19 machinery for stability and insertion in these membranes. This is supported by various data previously reported for S. cerevisiae and P. pastoris (Hettema et al., 2000; Hazra et al., 2002).
Interestingly, also in human cells Pex13 was shown to be able to insert into peroxisomal membranes independent of Pex19, whereas in yeast and mammals Pex13 is essential for the association of Pex14 with the peroxisomal membrane (Fransen et al., 2004). These data underscore that these two peroxins do not require the Pex3/Pex19 machinery for proper membrane insertion.
Conclusions
Peroxisome proliferation at inducing conditions is heavily debated but in yeast consensus is achieved that in these organisms fission is the main mode of organelle multiplication rather than de novo synthesis. Also in mammals the major mode of peroxisome proliferation is most likely fission, although de novo synthesis may occur as well.
De novo synthesis in yeast is observed in cells that lack peroxisomes. This process is in particular studied in pex3 strains upon reintroduction of the PEX3 gene. Recent data however indicated that pex3 cells contain peroxisomal vesicles that form in the absence of Pex3 (Knoops et al., 2014). It was shown that in pex3 cells not the ER but in fact these peroxisomal vesicles were the target for Pex3 and the subsequent formation of peroxisomes. However, various questions remain. For instance, it is unknown where the vesicles in pex3 cells originate from. They may arise by fission of existing structures but also form from the ER (Figure 2). In the latter view these in vivo data may complement recent in vitro studies in which pre-peroxisomes were formed from microsomal fractions (Lam et al., 2011) or in permeabilized cells (Agrawal et al., 2011). Clearly, the in vivo data suggest a novel concept of peroxisome reintroduction in pex3 cells and as such may promote future studies in this field. One approach may involve searching for novel proteins involved in de novo synthesis. A recent model that genetically separates de novo synthesis from fission may be useful in this respect. Two independent studies convincingly showed that mutants affected in fission or de novo synthesis do not display a peroxisome-deficient phenotype (Saraya et al., 2011; Huber et al., 2012). Only the combination of the two mutations as in a pex11 pex25 double mutant leads to the absence of peroxisomes. With this, an elegant screen is now available for identifying novel components involved in de novo synthesis by creating double mutants in a pex11 strain and select for peroxisome-deficient mutants.
Finally, the principles of PMP sorting are far from solved and change from the view that all PMPs travel via the ER to the assumption that PMPs travel directly to the target organelle. Considering the current literature, it is likely that both pathways in fact exist simultaneously. In this respect it is relevant to study the effect of manipulating modulation the affinity of the two sorting signals proposed (either for the ER or the intact peroxisome) for their substrate organelle. This may help in understanding why the protein travels to the ER in peroxisome-deficient mutants but to the intact organelle at WT conditions.
So far, most FM approaches used suffer from distinct drawbacks (i.e., overexpression effects) that do not allow drawing unequivocal conclusions for WT conditions. Clearly, novel techniques are required, like pulse chase experiments to show the transient ER location of specific PMPs in conjunctions with high speed microscopy techniques to track the routing of these proteins.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Rinse de Boer for preparing the figures.
References
Agrawal, G., Joshi, S., and Subramani, S. (2011). Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 108, 9113–9118. doi: 10.1073/pnas.1018749108
Cepinska, M. N., Veenhuis, M., van der Klei, I. J., and Nagotu, S. (2011). Peroxisome fission is associated with reorganization of specific membrane proteins. Traffic 12, 925–937. doi: 10.1111/j.1600-0854.2011.01198.x
Delille, H. K., Agricola, B., Guimaraes, S. C., Borta, H., Luers, G. H., Fransen, M., et al. (2010). Pex11pbeta-mediated growth and division of mammalian peroxisomes follows a maturation pathway. J. Cell Sci. 123, 2750–2762. doi: 10.1242/jcs.062109
Delille, H. K., and Schrader, M. (2008). Targeting of hFis1 to peroxisomes is mediated by Pex19p. J. Biol. Chem. 283, 31107–31115. doi: 10.1074/jbc.M803332200
Fagarasanu, A., Fagarasanu, M., Eitzen, G. A., Aitchison, J. D., and Rachubinski, R. A. (2006). The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev. Cell 10, 587–600. doi: 10.1016/j.devcel.2006.04.012
Fagarasanu, M., Fagarasanu, A., Tam, Y. Y., Aitchison, J. D., and Rachubinski, R. A. (2005). Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J. Cell Biol. 169, 765–775. doi: 10.1083/jcb.200503083
Fakieh, M. H., Drake, P. J., Lacey, J., Munck, J. M., Motley, A. M., and Hettema, E. H. (2013). Intra-ER sorting of the peroxisomal membrane protein Pex3 relies on its luminal domain. Biol. Open 2, 829–837. doi: 10.1242/bio.20134788
Fang, Y., Morrell, J. C., Jones, J. M., and Gould, S. J. (2004). PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J. Cell Biol. 164, 863–875. doi: 10.1083/jcb.200311131
Fransen, M., Vastiau, I., Brees, C., Brys, V., Mannaerts, G. P., and Van Veldhoven, P. P. (2004). Potential role for Pex19p in assembly of PTS-receptor docking complexes. J. Biol. Chem. 279, 12615–12624. doi: 10.1074/jbc.M304941200
Fujiki, Y., Rachubinski, R. A., Mortensen, R. M., and Lazarow, P. B. (1985). Synthesis of 3-ketoacyl-CoA thiolase of rat liver peroxisomes on free polyribosomes as a larger precursor. Induction of thiolase mRNA activity by clofibrate. Biochem. J. 226, 697–704.
Gandre-Babbe, S., and van der Bliek, A. M. (2008). The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 19, 2402–2412. doi: 10.1091/mbc.E07-12-1287
Goldman, B. M., and Blobel, G. (1978). Biogenesis of peroxisomes: intracellular site of synthesis of catalase and uricase. Proc. Natl. Acad. Sci. U.S.A. 75, 5066–5070. doi: 10.1073/pnas.75.10.5066
Haan, G. J., Baerends, R. J., Krikken, A. M., Otzen, M., Veenhuis, M., and van der Klei, I. J. (2006). Reassembly of peroxisomes in Hansenula polymorpha pex3 cells on reintroduction of Pex3p involves the nuclear envelope. FEMS Yeast Res. 6, 186–194. doi: 10.1111/j.1567-1364.2006.00037.x
Halbach, A., Landgraf, C., Lorenzen, S., Rosenkranz, K., Volkmer-Engert, R., Erdmann, R., et al. (2006). Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J. Cell Sci. 119, 2508–2517. doi: 10.1242/jcs.02979
Hazra, P. P., Suriapranata, I., Snyder, W. B., and Subramani, S. (2002). Peroxisome remnants in pex3delta cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic 3, 560–574. doi: 10.1034/j.1600-0854.2002.30806.x
Hettema, E. H., Girzalsky, W., van den Berg, M., Erdmann, R., and Distel, B. (2000). Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19, 223–233. doi: 10.1093/emboj/19.2.223
Hoepfner, D., Schildknegt, D., Braakman, I., Philippsen, P., and Tabak, H. F. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122, 85–95. doi: 10.1016/j.cell.2005.04.025
Hoepfner, D., van den Berg, M., Philippsen, P., Tabak, H. F., and Hettema, E. H. (2001). A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155, 979–990. doi: 10.1083/jcb.200107028
Huber, A., Koch, J., Kragler, F., Brocard, C., and Hartig, A. (2012). A subtle interplay between three Pex11 proteins shapes de novo formation and fission of peroxisomes. Traffic 13, 157–167. doi: 10.1111/j.1600-0854.2011.01290.x
Huber, N., Guimaraes, S., Schrader, M., Suter, U., and Niemann, A. (2013). Charcot-Marie-Tooth disease-associated mutants of GDAP1 dissociate its roles in peroxisomal and mitochondrial fission. EMBO Rep. 14, 545–552. doi: 10.1038/embor.2013.56
Huybrechts, S. J., Van Veldhoven, P. P., Brees, C., Mannaerts, G. P., Los, G. V., and Fransen, M. (2009). Peroxisome dynamics in cultured mammalian cells. Traffic 10, 1722–1733. doi: 10.1111/j.1600-0854.2009.00970.x
Kal, A. J., van Zonneveld, A. J., Benes, V., van den Berg, M., Koerkamp, M. G., Albermann, K., et al. (1999). Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol. Biol. Cell 10, 1859–1872. doi: 10.1091/mbc.10.6.1859
Kiel, J. A., Veenhuis, M., and van der Klei, I. J. (2006). PEX genes in fungal genomes: common, rare or redundant. Traffic 7, 1291–1303. doi: 10.1111/j.1600-0854.2006.00479.x
Kim, P. K., Mullen, R. T., Schumann, U., and Lippincott-Schwartz, J. (2006). The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173, 521–532. doi: 10.1083/jcb.200601036
Knoblach, B., Sun, X., Coquelle, N., Fagarasanu, A., Poirier, R. L., and Rachubinski, R. A. (2013). An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 32, 2439–2453. doi: 10.1038/emboj.2013.170
Knoops, K., Manivannan, S., Cepińska, M. N., Krikken, A. M., Kram, A. M., Veenhuis, M., et al. (2014). Preperoxisomal vesicles can form in the absence of Pex3. J. Cell Biol. 204, 659–668. doi: 10.1083/jcb.201310148
Koch, A., Thiemann, M., Grabenbauer, M., Yoon, Y., Mcniven, M. A., and Schrader, M. (2003). Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278, 8597–8605. doi: 10.1074/jbc.M211761200
Kragt, A., Voorn-Brouwer, T., van den Berg, M., and Distel, B. (2005). Endoplasmic reticulum-directed Pex3p routes to peroxisomes and restores peroxisome formation in a Saccharomyces cerevisiae pex3Delta strain. J. Biol. Chem. 280, 34350–34357. doi: 10.1074/jbc.M505432200
Kuravi, K., Nagotu, S., Krikken, A. M., Sjollema, K., Deckers, M., Erdmann, R., et al. (2006). Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119, 3994–4001. doi: 10.1242/jcs.03166
Lam, S. K., Yoda, N., and Schekman, R. (2011). A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 108, E51–E52. doi: 10.1073/pnas.1103526108
Lazarow, P. B., and Fujiki, Y. (1985). Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1, 489–530. doi: 10.1146/annurev.cb.01.110185.002421
Matsuzaki, T., and Fujiki, Y. (2008). The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J. Cell Biol. 183, 1275–1286. doi: 10.1083/jcb.200806062
Matsuzono, Y., and Fujiki, Y. (2006). In vitro transport of membrane proteins to peroxisomes by shuttling receptor Pex19p. J. Biol. Chem. 281, 36–42. doi: 10.1074/jbc.M509819200
Menendez-Benito, V., van Deventer, S. J., Jimenez-Garcia, V., Roy-Luzarraga, M., van Leeuwen, F., and Neefjes, J. (2013). Spatiotemporal analysis of organelle and macromolecular complex inheritance. Proc. Natl. Acad. Sci. U.S.A. 110, 175–180. doi: 10.1073/pnas.1207424110
Motley, A. M., and Hettema, E. H. (2007). Yeast peroxisomes multiply by growth and division. J. Cell Biol. 178, 399–410. doi: 10.1083/jcb.200702167
Motley, A. M., Ward, G. P., and Hettema, E. H. (2008). Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell Sci. 121, 1633–1640. doi: 10.1242/jcs.026344
Nagotu, S., Krikken, A. M., Otzen, M., Kiel, J. A., Veenhuis, M., and van der Klei, I. J. (2008a). Peroxisome fission in Hansenula polymorpha requires Mdv1 and Fis1, two proteins also involved in mitochondrial fission. Traffic 9, 1471–1484. doi: 10.1111/j.1600-0854.2008.00772.x
Nagotu, S., Saraya, R., Otzen, M., Veenhuis, M., and van der Klei, I. J. (2008b). Peroxisome proliferation in Hansenula polymorpha requires Dnm1p which mediates fission but not de novo formation. Biochim. Biophys. Acta 1783, 760–769. doi: 10.1016/j.bbamcr.2007.10.018
Opalinski, L., Kiel, J. A., Williams, C., Veenhuis, M., and van der Klei, I. J. (2011). Membrane curvature during peroxisome fission requires Pex11. EMBO J. 30, 5–16. doi: 10.1038/emboj.2010.299
Otzen, M., Ruckt, R., Thoms, S., Emmrich, K., Krikken, A. M., Erdmann, R., et al. (2012). Pex19p contributes to peroxisome inheritance in the association of peroxisomes to Myo2p. Traffic 13, 947–959. doi: 10.1111/j.1600-0854.2012.01364.x
Pinto, M. P., Grou, C. P., Alencastre, I. S., Oliveira, M. E., Sa-Miranda, C., Fransen, M., et al. (2006). The import competence of a peroxisomal membrane protein is determined by Pex19p before the docking step. J. Biol. Chem. 281, 34492–34502. doi: 10.1074/jbc.M607183200
Raychaudhuri, S., and Prinz, W. A. (2008). Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 105, 15785–15790. doi: 10.1073/pnas.0808321105
Rucktaschel, R., Halbach, A., Girzalsky, W., Rottensteiner, H., and Erdmann, R. (2010). De novo synthesis of peroxisomes upon mitochondrial targeting of Pex3p. Eur. J. Cell Biol. 89, 947–954. doi: 10.1016/j.ejcb.2010.06.012
Saraya, R., Krikken, A. M., Veenhuis, M., and van der Klei, I. J. (2011). Peroxisome reintroduction in Hansenula polymorpha requires Pex25 and Rho1. J. Cell Biol. 193, 885–900. doi: 10.1083/jcb.201012083
Schliebs, W., and Kunau, W. H. (2004). Peroxisome membrane biogenesis: the stage is set. Curr. Biol. 14, R397–R399. doi: 10.1016/j.cub.2004.05.017
Schuldiner, M., Metz, J., Schmid, V., Denic, V., Rakwalska, M., Schmitt, H. D., et al. (2008). The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634–645. doi: 10.1016/j.cell.2008.06.025
South, S. T., Baumgart, E., and Gould, S. J. (2001). Inactivation of the endoplasmic reticulum protein translocation factor, Sec61p, or its homolog, Ssh1p, does not affect peroxisome biogenesis. Proc. Natl. Acad. Sci. U.S.A. 98, 12027–12031. doi: 10.1073/pnas.221289498
Tam, Y. Y., Fagarasanu, A., Fagarasanu, M., and Rachubinski, R. A. (2005). Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J. Biol. Chem. 280, 34933–34939. doi: 10.1074/jbc.M506208200
Thoms, S., Harms, I., Kalies, K. U., and Gartner, J. (2012). Peroxisome formation requires the endoplasmic reticulum channel protein Sec61. Traffic 13, 599–609. doi: 10.1111/j.1600-0854.2011.01324.x
Titorenko, V. I., Chan, H., and Rachubinski, R. A. (2000). Fusion of small peroxisomal vesicles in vitro reconstructs an early step in the in vivo multistep peroxisome assembly pathway of Yarrowia lipolytica. J. Cell Biol. 148, 29–44. doi: 10.1083/jcb.148.1.29
van der Zand, A., Braakman, I., and Tabak, H. F. (2010). Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol. Biol. Cell 21, 2057–2065. doi: 10.1091/mbc.E10-02-0082
van der Zand, A., Gent, J., Braakman, I., and Tabak, H. F. (2012). Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149, 397–409. doi: 10.1016/j.cell.2012.01.054
Veenhuis, M., van Dijken, J. P., Pilon, S. A., and Harder, W. (1978). Development of crystalline peroxisomes in methanol-grown cells of the yeast Hansenula polymorpha and its relation to environmental conditions. Arch. Microbiol. 117, 153–163. doi: 10.1007/BF00402303
Waterham, H. R., Koster, J., van Roermund, C. W., Mooyer, P. A., Wanders, R. J., and Leonard, J. V. (2007). A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 356, 1736–1741. doi: 10.1056/NEJMoa064436
Wriessnegger, T., Gubitz, G., Leitner, E., Ingolic, E., Cregg, J., De La Cruz, B. J., et al. (2007). Lipid composition of peroxisomes from the yeast Pichia pastoris grown on different carbon sources. Biochim. Biophys. Acta 1771, 455–461. doi: 10.1016/j.bbalip.2007.01.004
Yagita, Y., Hiromasa, T., and Fujiki, Y. (2013). Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway. J. Cell Biol. 200, 651–666. doi: 10.1083/jcb.201211077
Keywords: peroxisome, fission, endoplasmic reticulum, de novo peroxisome formation, yeast
Citation: Veenhuis M and van der Klei IJ (2014) A critical reflection on the principles of peroxisome formation in yeast. Front. Physiol. 5:110. doi: 10.3389/fphys.2014.00110
Received: 15 December 2013; Paper pending published: 17 February 2014;
Accepted: 04 March 2014; Published online: 20 March 2014.
Edited by:
Vladimir I. Titorenko, Concordia University, CanadaReviewed by:
Vladimir I. Titorenko, Concordia University, CanadaMarc Fransen, Katholieke Universiteit Leuven, Belgium
Copyright © 2014 Veenhuis and van der Klei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ida J. van der Klei, Molecular Cell Biology, University of Groningen, PO Box 11103, 9700 CC Groningen, Netherlands e-mail: i.j.van.der.klei@rug.nl
 Marten Veenhuis
Marten Veenhuis Ida J. van der Klei
Ida J. van der Klei