- 1Department of Biological Sciences, George Washington University, Washington, DC, United States
- 2Department of Biological Sciences, University of Alberta, Edmonton, AB, Canada
Overcrowding conditions and temperatures shifts regularly manifest in large-scale infections of farmed fish, resulting in economic losses for the global aquaculture industries. Increased understanding of the functional mechanisms of fish antimicrobial host defenses is an important step forward in prevention of pathogen-induced morbidity and mortality in aquaculture setting. Like other vertebrates, macrophage-lineage cells are integral to fish immune responses and for this reason, much of the recent fish immunology research has focused on fish macrophage biology. These studies have revealed notable similarities as well as striking differences in the molecular strategies by which fish and higher vertebrates control their respective macrophage polarization and functionality. In this review, we address the current understanding of the biological mechanisms of teleost macrophage functional heterogeneity and immunity, focusing on the key cytokine regulators that control fish macrophage development and their antimicrobial armamentarium.
Introduction
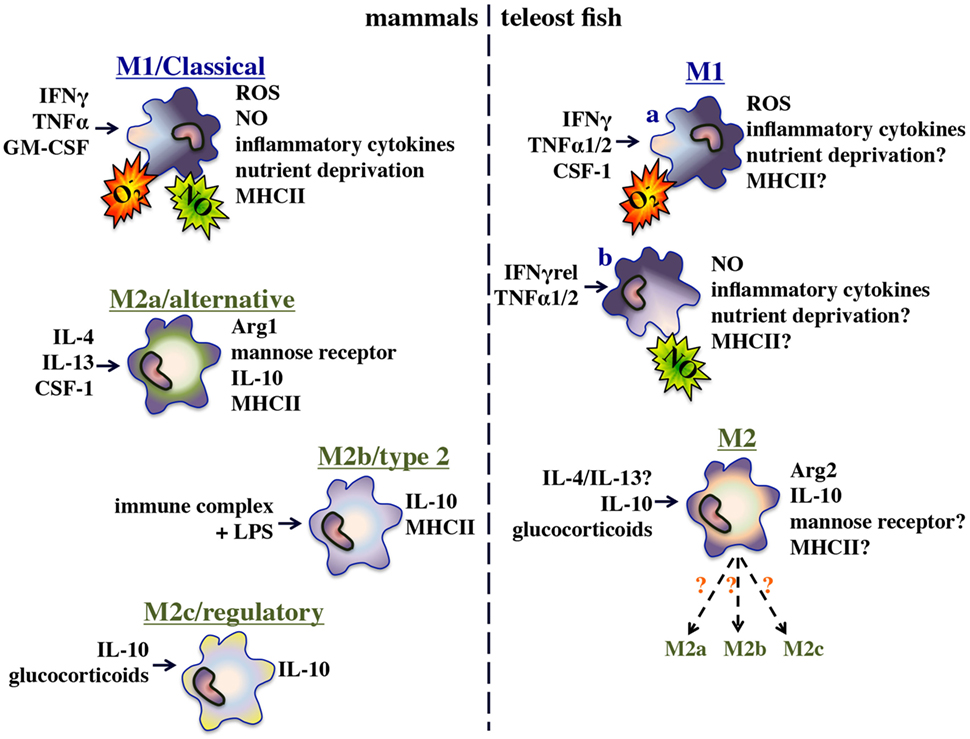
The immune systems of all vertebrates are integrally dependent on macrophage-lineage cells and while the ontogeny of functionally disparate macrophage subsets and lineages have been thoroughly studied in mammals (1, 2), they remain to be adequately defined in aquatic vertebrates such as teleost fish [previously reviewed by Hodgkinson et al. (3)]. In mammals, these functionally distinct macrophage subsets are framed by polarized extremes including the interferon-gamma (IFNγ) and tumor necrosis factor-alpha (TNFα) primed M1/classically activated macrophages; the interleukin-4 and/or interleukin-13-stimulated M2a/alternatively activated macrophages; the immune complexes or apoptotic cell-stimulated M2b/alternatively activated macrophages; and the interluekin-10 (IL-10), transforming growth factor-beta (TGF-β) and/or glucocorticoid (GC)-primed M2c/alternatively polarized macrophages (4). Depending on their respective stimulus-dependent polarization states, these macrophage subsets participate in either inflammatory/microbicidal or repair/wound-healing/immune suppression responses (4). While bony fish clearly possess functional analogs of these mammalian macrophage subsets (Figure 1), the molecular mechanisms governing the polarization and functionality of these respective fish macrophage populations remain to be fully defined.
The teleost fish inflammatory/M1 macrophage populations have been the best-studied and shown to rapidly kill pathogens through phagocytosis (5), production of reactive oxygen and nitrogen intermediates (6, 7), and restriction of nutrient availability (8, 9). Furthermore and akin to their mammalian counterparts, these fish M1 macrophages produce a plethora of inflammatory cytokines, chemokines, and lipid mediators (9). In a recent effort to gain insights into the alternatively polarized/M2 fish macrophages, researchers have examined functional macrophage parameters such as arginase gene expression and activity (10) and their expression of immunosuppressive cytokines such as il10 and tgfβ (11–13). Indeed, the functional analogs of the mammalian M1/M2a–c macrophage subsets appear to be present in teleosts. However, defining the regulatory mechanisms governing the polarization of these effector populations is a far more challenging goal as gene-specific and whole genome duplication events have endowed disparate fish species with unique multi-copy repertoires of those genes, which in mammals are though to dictate macrophage polarization and functionality (14).
In this review, we focus on the current understanding of the molecular mechanisms of fish macrophage antimicrobial responses to prokaryotic and eukaryotic pathogens.
Macrophage Ontogeny
Macrophage Sources and Fates
Until recently, tissues macrophages were believed to arise from circulating monocyte precursors in response to tissue entry and accompanying stimuli (15). However, more recent research has challenged this notion and suggests that while mammalian blood monocytes may enter into tissues and become macrophages under certain inflammatory conditions, these events are infrequent (15–17). Instead, mammalian resident tissue macrophages are now thought to be seeded during embryonic hematopoiesis and replenish resident populations locally (16–18). While the presence of self-renewing fish tissue macrophage populations requires further investigation (19), recent reports showed that fish lacking functional c-myb transcriptional regulator of adult hematopoiesis, nonetheless possess tissue macrophages suggest that this process may be conserved in teleosts (20).
Teleost Monopoiesis and the Colony-Stimulating Factor-1 Receptor (CSF-1R)
The differentiation and functionality of most vertebrate macrophages are controlled by engagement of the CSF-1R, which is expressed on committed myeloid precursor cells and their derivative populations (21–23). The csf1r (fms) genes of different vertebrate species exhibit poor sequence identities, particularly in their extracellular domains (24–26). By contrast, the catalytic tyrosine kinase domains of CSF-1Rs are highly conserved (27, 28). The divergence of the extracellular portions of the CSF-1R molecules likely reflects the selective pressure onto this receptor of diverging (and in some cases multiple) ligands of these receptors, as these exhibit low amino acid sequence conservation. The mammalian, reptilian, avian, and teleost fish CSF-1Rs all branch into phylogenetically separate clades (26), presumably reflecting the many distinct aspects of macrophage functionality across these divergent species. In turn, these differences may reflect distinct functional contributions of these respective ligands and receptor systems to the macrophage ontogeny and functionality of the evolutionarily diverged vertebrate species.
Colony-Stimulating Factor-1
Unlike birds and mammals that have a single alternatively spliced csf1 gene (29, 30), many teleost fish species have two distinct csf1 genes (csf1.1 and csf1.2), which (for the most part) do not appear to undergo alternative splicing (25). Like its mammalian counterpart, the fish CSF-1 (CSF-1.1) also appears to be an important macrophage growth and differentiation factor (31, 32). Interestingly, while the mammalian CSF-1 is known for driving alternative/M2 macrophage differentiation (28), the cyprinid (goldfish) CSF-1.1 appears to facilitate the functional differentiation of inflammatory/M1-like macrophages with highly upregulated pro-inflammatory components (32). This is supported by the reports that a soluble goldfish CSF-1R (19) down-regulates macrophage pro-inflammatory responses by reducing available soluble CSF-1 (33, 34) (see section below). As teleosts possess multiple csf1 genes and at least some fish species also encode two distinct csf1r genes (35), this suggests that teleost fish may have adopted more elaborate macrophage differentiation strategies to those seen in mammals.
Interleukin-34 (IL-34) as Possible Sources of Macrophage Functional Heterogeneity
Inflammatory (M1) macrophages produce multiple inflammatory mediators that coordinate antimicrobial responses, while the alternatively activated (M2) macrophages secrete immunosuppressive and angiogenic compounds that control the resolution of inflammation [reviewed by Zhou et al. (4) and Hodgkinson et al. (3)]. The mammalian CSF-1 induces the differentiation/polarization of M2 macrophages (28), whereas the teleost CSF-1 elicits an M1-like macrophage phenotype (32). Notably, the IL-34 cytokine also ligates and activates the CSF-1R (36–38), regulating the development of mammalian osteoclasts (39, 40), Langerhans cells (41, 42), microglia (41), and B cell-stimulating myeloid cells (43). Recent work using the amphibian Xenopus laevis model indicated that frog macrophages differentiated by the X. laevis CSF-1 are highly susceptible to the emerging Frog Virus 3 Ranavirus whereas macrophages derived by IL-34 are important antiviral effectors (26, 44, 45). The antiviral roles of IL-34-derived macrophages remain to be fully elucidated in other vertebrates, and it is likely that akin to CSF-1, IL-34 likewise contributes to macrophage functional heterogeneity.
To date, there have been a limited number of studies addressing the contribution of IL-34 to the fish macrophage biology. Recent work indicates that the grouper IL-34 plays an important role in the fish immune response against Cryptocaryon irritans infections, as the expression of this gene was highly upregulated in the parasite-infected fish gill and skin tissues (46). This is consistent with the roles of the mammalian IL-34 in the differentiation and functionality of tissue resident macrophages and Langerhans cells (41, 42) and may reflect an evolutionarily conserved role for IL-34 in controlling the development of this macrophage-lineage cell type.
The trout il34, csf1.1, and csf1.2 are differentially expressed in fish tissues and as well as in a number of trout-derived cell lines, suggesting disparate biological roles for these CSF-1R ligands (47). Notably, the trout il34 exhibited high baseline tissue expression in which the authors attributes to a possible homeostatic role and that indeed could reflect the conserved role of this growth factor in tissue macrophage and Langerhans cell biology. Moreover, whereas stimulation of primary trout kidney macrophage cultures with a number of pathogen-associated molecular patterns (PAMPs) failed to elicit increases in csf1.1 or csf1.2 gene expression, these stimuli readily upregulated the expression of il34 by these cells (47). Notably, the viral dsRNA mimic poly I:C elicited a particularly robust increase the macrophage il34 expression, possibly reflecting a conserved role for the fish IL-34 in antiviral immunity, akin to the amphibian counterpart.
Soluble CSF-1R
Cyprinid fish control their CSF-1 (and presumably IL-34) stimulation of macrophages by production of a soluble CSF-1 receptor (sCSF-1R) (33, 34, 48, 49). This soluble form of the receptor arises through alternative splicing, and is capable of ablating macrophage proliferation (48) and macrophage-mediated inflammatory responses (33, 49). The sCSF-1R is produced by mature macrophages, but not monocytes, in response to classical M2-polarizing stimuli such as apoptotic cells (34) and efficiently ablates an array of inflammatory events including leukocyte infiltration (34), macrophage chemotaxis, phagocytosis, production of reactive oxygen intermediates and recruitment of leukocytes (33). Moreover, sCSF-1R dampens fish macrophage chemokine and inflammatory cytokine expression, neutrophil recruitment while promoting the expression of the anti-inflammatory cytokine, interleukin-10 (49). It will be interesting to learn whether other fish besides cyprinids have adopted this strategy for controlling their macrophage inflammatory responses.
Molecular Control of Macrophage Antimicrobial Armamentarium
Pattern Recognition Receptors (PRRs) of Teleost Macrophages
During injury and/or infection, resident macrophages detect tissue damage and/or infiltrating pathogens by either extracellular or intracellular pattern recognition receptors (PRRs). The existence of immune PRRs was first proposed by Charles Janeway over 20 years ago (50). The known PRRs can be classified into five groups based on their structure and function: toll-like receptors (TLRs), C-type lectins, nucleotide-binding domain-leucine-rich repeat containing receptors (NLRs or NOD-like), retinoic acid inducible gene 1 (RIG1)-like receptors (RLRs), and absence in melanoma (AIM)-like receptors (ALRs) [reviewed by Hansen et al. (51)]. Neutrophils, monocytes, macrophages, dendritic cells (DCs), and specific epithelial and endothelial cells have PRRs (51).
In addition to specific recognition of distinct pathogen components (e.g., LPS, dsRNA, and flagellin), PRRs also detect tissue damage-associated molecular patterns (52–54). The human TLRs 1, 2, 4, 5, 6, and 10 are membrane bound while the TLRs 3, 7, 8, and 9 are located in endosomes (55). By contrast, the NLRs, RLRs, and ALRs are exclusively cytosolic (55).
Members of the TLR family share the intracellular toll-interleukin-1 receptor motifs (56). Initially identified in Drosophila spp. for controlling dorso-ventral patterning (57) and subsequently attributed to its anti-fungal properties (58), members of this family are now widely believed to be indispensable for immune recognition by most metazoans. Humans are currently known to have 10 TLRs (TLR 1–10) and mice possess 12 TLRs (51, 59). Birds also possess 10 TLRs, of which some are counterparts of the mammalian receptors (TLR 3–5, 7, and two forms of each TLR 1 and 2) (59). Some birds (TLR15, 16, and 21) are not found in higher vertebrates (60), and amphibians may have up to 20 TLRs (61). Bony fish possess 17 distinct TLRs, including some that are unique to fish, such as TLR 20–23 (62–64). Interestingly, not all fish have TLR4 and zebrafish TLR4 does not recognize LPS and negatively regulates NF-kB signaling (51, 65, 66). Additional research will be required to fully elucidate the function of TLRs in lower vertebrates, which will undoubtedly shed new light on the evolutionary history of these important innate immune receptors. The biology of fish TLRs has recently been a subject to several excellent review articles (67–70).
Other PRR families have also been identified in aquatic vertebrates. Although, gene synteny analyses identified a number of RLRs in birds and fish (71, 72), certain RLR genes are either absent or diverged beyond recognition. The evidence of functional conservation of fish RLRs exists (73, 74).
The NLRs were originally discovered in plants as R-proteins, which share nucleotide binding site and leucine rich repeat domains and can detect proteins delivered by pathogenic bacteria to trigger rapid activation of host defense (75, 76). The first identified mammalian NLR was the human NOD1 (also known as CARD4) by Bertin et al. (77) and Inohara et al. (78). The NOD1/CARD4 contained the typical NOD domain (also referred as the NACHT domain), which is a critical structural feature of NLRs (79–81), and NOD2/Card15 was identified searching for NOD1 homologs in genomic databases (82), and at present there are 23–34 NLRs known to exist in humans and mice, respectively.
There are several orthologs of mammalian NLRs as well as a unique NLR subfamily of receptors in bony fish (83). The first reported teleost NLRs were identified in zebrafish genome (84). Three subfamilies of NLRs were present in zebrafish, the first resembled mammalian NODs, the second resembled mammalian NLRPs, and the third was reported to be a unique subfamily of genes having similarities to both mammalian NOD3 and NLRPs (83). The existence of NLRs has been reported in grass carp (85), rainbow trout (86), channel catfish (87, 88), common carp (89, 90), orange-spotted grouper (91), goldfish (92), Japanese founder (93, 94), miiuy croaker (95, 96), and Japanese pufferfish (97). The results of these studies indicated the presence of inducible NLRs and that teleost NLRs shared the conserved structural domains with their mammalian counterparts. Studies on most of teleost macrophage NLRs primarily focused on the examination of gene expression induced by different immune stimuli and/or fish pathogens (87, 88, 92) and to a lesser extent on NLR signaling pathways in fish macrophages (98–102).
The Type II Interferon System(s) of Bony Fish
The classical/M1 macrophage activation corresponds to macrophage upregulation of an array of inflammatory, microbicidal, and antigen presentation components, and is linked due to Th1-biased cytokine stimulation of these cells (103, 104). Specifically, this classical macrophage activation is thought to predominantly occur in response to the type II interferon cytokine, IFNγ, which is produced by Th1 helper cells and activated NK cells (105, 106). The induction of the mammalian M1 macrophages requires the co-stimulation of cells with IFNγ and TNFα (107). Conversely, these classically activated macrophages may be generated following macrophage activation through pathogen PRRs (108). While teleost fish have numerous PRRs (83, 109), the roles of fish PRRs (see previous section) in teleost M1 macrophage polarization remains to be fully addressed.
The mammalian IFNγ cytokine has been linked to an vast array of immunological processes, and was first identified from the supernatants of PHA-activated lymphocytes (110). In addition to its modest antiviral capacities, IFNγ appears to be particularly important to vertebrate host defenses against obligate and facultative intracellular pathogens (111–115). These include several important macrophage pathogens such as Listeria monocytogenes (116), Leishmania major (117), and Mycobacterium (118). This underlines the importance of this cytokine to macrophage immunity (111, 119–122).
The mammalian IFNγ binds the interferon gamma receptor 1 (IFNGR1), which results in the formation of a receptor complex composed of this ligand binding chain as well as the IFNGR2 signal propagation chain, ensuing in the downstream signaling cascade (123). The assembly of this signaling complex (IFNγ:IFNGR1:IFNGR2) activates Janus kinases (Jak)-1 and -2 (124), upon which phosphorylation activates signal transducer of activation-1 (Stat1) transcription factor (125). Under certain cellular conditions, stimulation with IFNγ may also activate Stat2 (126) albeit to a much lesser extent than Stat1. Moreover, IFNγ signaling typically results in the activation and nuclear translocation of several other transcriptional complexes including ISGF3 and Stat1-p48, composed of Stat1: Stat2: IRF-9 and Stat1: Stat1: IRF-9 (126–129). IFNγ signaling occurs in temporal phases, where the first sets of interferon gamma stimulated transcripts are seen after 30 min of the initial IFNγ receptor activation, and many of the products of these mRNAs then modulate subsequent IFNγ-related (IFNγrel) signaling events within the stimulated cell (130).
Teleost fish are widely known to possess ifng genes (131–135) and the functional roles attributed to the mammalian IFNγs appear to be conserved to these fish cytokine counterparts. For example, the trout IFNγ elicits the expression of a number of immune genes such as γip10, mhcIIb, and stat1 (136), c-type lectin, il1b, ifng, tap1, tapasin, irf1, ikb, and junb in the monocyte/macrophage RTS11 cell line (137). Fish IFNγ enhances reactive oxygen species (ROS) production by primary kidney phagocytes of trout (136), goldfish (138), and carp (139). The goldfish IFNγ primes kidney-derived monocyte ROS responses in a concentration dependent manner (138) and akin to its mammalian counterpart (140, 141), the goldfish IFNγ synergizes with the goldfish TNFα (138) to prime the fish monocyte ROS response. The goldfish IFNγ also induces modest but significant increases in kidney macrophage nitric oxide responses, which are further enhanced by co-stimulation with TNFα2 (138). Interestingly, the carp IFNγ elicits significant NO responses in fish kidney phagocytes only in conjunction with a high dose of LPS (139). The large yellow croaker IFNγ enhances the primary kidney phagocyte respiratory burst and nitric oxide responses and upregulates the gene expression of inflammatory genes such as tnfa, il1b, stat1, and irf1 in these cells (142). Likewise, the black seabream and the zebrafish IFNγs induce the expression of jaks, stats, and interferon-stimulated genes such as irf1 and mx (143, 144).
Goldfish kidney-derived macrophages stimulated with IFNγ upregulate their expression of several inflammatory genes including tnfa isoforms 1&2, il1b isoforms 1&2, il12 subunits p35 & p40, ifng, il8 (CXCL-8), ccl1, and viperin (138). Carp kidney phagocytes treated with IFNγ and LPS increase their gene expression of tnfa, il1b, and il12; (subunits p35 & p40) (139). Carp IFNγ also induced the expression of a CXCL-10-like chemokine (cxclb) and inhibited LPS-induced expression of cxcl8 (139). Together, it would appear that for the most part, the inflammatory roles of IFNγ such as its synergism with LPS and TNFα (see below) are conserved in teleosts.
Functional Dichotomy of Fish Type II IFNs
In fish, Igawa et al. (132) identified two genes encode ifng isoforms, located next to the fish il22 and il26 genes, that have the exon/intron organization of ifng genes of other vertebrates and possess the IFNγ signature motif ([IV]-Q-X-[KQ]-A-X2-E-[LF]-X2-[IV]). These two ifng sequences were initially coined IFNγ1 and IFNγ2 but following a reevaluation of vertebrate ifng genes and because the fish IFNγ2 possesses the hallmark features of the mammalian IFNγs, it was renamed as simply IFNγ (145). Since IFNγ1 appears to be structurally related to the mammalian IFNγ, but is missing a nuclear localization signal (NLS) motif, it has been coined as IFNγrel. The presence of multiple ifng isoforms have now been confirmed in siluriformes and other cypriniformes, including the identification of ifngrel and ifng in catfish (133), common carp (146), zebrafish (147), and the goldfish (138).
The siluriform IFNγrel proteins have not been functionally characterized. However, the cyprinid IFNγrels have been examined in some detail across several species. For example, freshly laid zebrafish eggs possess ifngrel transcripts, indicating maternal supply of these mRNAs (147). Also, while the gene expression of the zebrafish ifng is not detected until much later in development, the mRNA levels of IFNγrel continue to increase during the embryonic zebrafish development (147). Moreover, injection of zebrafish embryos with mRNAs encoding IFNγ or IFNγrel results in increased expression of genes typically activated by the mammalian IFNγ (147). Notably, morpholino knock-down of either ifng or ifngrel resulted in compromised Yersinia ruckeri-infected zebrafish embryo survival while the combined knock-down of both cytokines further decreased embryo survival (147), suggesting that IFNγ or IFNγrel confers at least partially non-overlapping immune roles.
The goldfish IFNγ and IFNγrel appear to confer distinct effects on macrophages (138, 148). For example, while IFNγ stimulation of goldfish monocytes results in long-lasting ROS priming, IFNγrel elicits a short-lived priming effect on these cells, followed by complete monocyte unresponsiveness to ROS priming by other inflammatory cytokines (IFNγ or TNFα2). Moreover, the goldfish IFNγ only modestly enhances fish monocyte/macrophage phagocytosis and nitric oxide responses (138, 148). By stark contrast, IFNγrel induced significantly greater phagocytosis, iNOS (isoforms A and B) gene expression, and nitric oxide production in goldfish monocytes and macrophages. Interestingly, these goldfish type II IFNs also elicit the expression of distinct immune genes in goldfish monocytes. Both recombinant cytokines induce goldfish monocyte Stat1 phosphorylation, however, nuclear translocation of Stat1 was only seen in cells treated with IFNγ, but not with IFNγrel. This was confirmed by more recent report, indicating that the zebrafish IFNγ and IFNγrel utilize distinct signaling pathways (143). It is interesting that while the recombinant ginbuna crucian carp IFNγ forms a dimer in solution, the recombinant IFNγrel appears to be monomeric (149, 150), akin to the functional forms of type I rather than type II IFNs. Moreover, an additional isoform of the ginbuna carp IFNγrel has been identified and shown to possess a functional NLS, which contrasts the other fish IFNγrel proteins (150). With the growing evidence indicating functional dichotomies of the cyprinid type II IFNs, it will be interesting to learn the roles of these distinct macrophage-activating factors in their target cells’ antimicrobial responses to different fish pathogens.
Fish Type II IFN Receptors
While the bony fish type II IFN ligands have become a subject of active research, the functional roles of the type II IFN receptors remain to be clearly defined. The trout IFNGR1 and IFNGR2 chains were initially identified and shown to exhibit conserved gene synteny across vertebrates (151). All fish IFNGR1 sequences have Jak1 and Stat1 binding sites, that are also required for functional mammalian IFNγ (152–154), and the expression of the IFNGR2 chain appears to be essential to the trout IFNγ-induced signaling (151).
The fish IFNγ and IFNγrel cytokines structural, functional, and intracellular signaling differences were thought to reflect the presence of distinct IFNγ receptors, dedicated to these respective moieties. As predicted, gene synteny analyses of the vertebrate ifngr1 genes (encoding the ligand binding chain), revealed two distinct zebrafish ifngr1 genes, located on distinct chromosomes (155). The presence of corresponding ifngr isoforms was confirmed in goldfish, and by means of in vitro recombinant protein binding studies, we demonstrated that IFNγrel (IFNγ1) and IFNγ each bound to their own cognate IFNγ receptor chains, the IFNGR1-1 and IFNGR1-2, respectively. Morpholino knock-down of the zebrafish ifngr1-1, ifngr1-2, or ifngr2 (signal propagation chain) abolished the fish IFNγ function (156). Notably, only the knock-down of ifngr1-1, but not ifngr1-2 or ifngr2, abrogated IFNγrel stimulation, suggesting that zebrafish IFNγ signals through a heterodimer (IFNGR1-1 and IFNGR1-2) and a IFNGR2 homodimer whereas the IFNγrel binds to homo-dimeric IFNGR1-1 and a distinct unknown receptor 2 chain. The discrepancy between these finds and our studies, which indicated IFNγ-IFNGR1-2 but not IFNγ-IFNGR1-1 interactions, could be explained in several ways. Aside from the possible species-specific differences, it may be that IFNGR1-1 binds IFNγ with lower affinity, explaining our inability top detect this interaction in vitro by western blot. Conversely, the presence of ifngrel mRNA in fresh zebrafish embryos (147), suggests that this cytokine may plays roles during zebrafish (and presumably other cyprinid fish) development. If is the case, morpholino knock down of its cognate receptor encoding gene, ifngr1-1 may manifest in reduced IFNγ function, as an indirect consequence of the abrogated IFNγrel-mediated immune development rather than through direct IFNγ–IFNGR1-1 interactions.
It is notable that using HeLa cells transfected with the ginbuna carp ifngr1-1 and ifngr1-2 encoding plasmids, it was shown that the carp IFNγ isoform 1 exclusively signals through the IFNGR1-2 whereas the IFNγ isoform 2 signals through the IFNGR1-1 (149). It is well established that both the mammalian and fish IFNγ signaling requires IFNGR2 chains (123, 156) while the fish Jak and Stat proteins have significantly diverged from (and are present in multiple forms as compared to) the mammalian counterparts (157).
While all other vertebrates examined to date encode individual type II IFNs and IFNGR1 genes, it is intriguing that certain fish possess two distinct IFN gamma-receptor binding chains (IFNGR1-1 and IFNGR1-2) as well as multiple type II IFNs (148, 149, 156). This suggests that these fish have adopted very unique strategies surrounding their principal M1 macrophage-activating cytokine system(s) and it will be exciting to learn what are the functional consequences of these differences.
Teleost TNFα
The mammalian TNFα is involved in a broad array of immunological roles (158–160). The name of this cytokine stems from its discovery in tumoricidal sera of Bacillus Calmette-Guerin-primed, endotoxin-treated mice (161). During vertebrate inflammatory response, TNFα promotes the chemotaxis of neutrophils and monocytes/macrophages to the sites of inflammation (162, 163), enhance macrophage phagocytosis (164–166), primes reactive oxygen and reactive nitrogen responses (167, 168), facilitates the chemotaxis of fibroblasts (169) and the release of platelet activating factors (170–172). Mammalian TNFα confers its immune effects either as a 17 kDa soluble protein or a 26 kDa type II trans-membrane protein (173–175) and most effects are induced after binding of homotrimerized TNFα to either the TNF-R1 or TNF-R2 (176, 177).
Tumor necrosis factor-alpha orthologs, possessing the TNF family signature [LV]-x-[LIVM]-x3-G-[LIVMF]-Y-[LIMVMFY]2-x2-[QEKHL] have been identified in several teleosts (178), underlining the evolutionary conservation of this cytokine. Like its mammalian counterpart, the teleost fish TNFα is a reliable marker of fish M1 macrophages (179, 180). Most fish species possess multiple TNFα isoforms (178, 181–188). These TNFα isoforms confer pro-inflammatory effects such as enhancing inflammatory gene expression, macrophage chemotaxis and phagocytosis, and eliciting phagocyte reactive oxygen and nitrogen intermediate production (183–185, 189–194). The in vivo roles of TNFα during fish inflammatory and M1 macrophage immune responses have also been confirmed in zebrafish (179), sole (195) and trout (196, 197).
Teleost TNFα Receptors
Bobe and Goetz (198) were first to report the presence of a death domain-containing TNF receptor in zebrafish and coined this gene the ovarian TNF receptor (otr), while putative zebrafish tnfr1 and tnfr2 gene sequences were deposited to GenBank, with the zebrafish tnfr1 sharing high sequence identity with otr. We identified the goldfish tnfr1 and tnfr2 cDNAs (199) and showed that the putative amino acid sequences of these goldfish receptors share many conserved regions with their respective mammalian counterparts. Goldfish TNF-R1 has a death domain with a conserved motif (W/E)-X31-L-X2-W-X12-L-X3-L and six residues that are essential to TNF-R1-mediated cytotoxicity (200).
Our in vitro binding studies using recombinant version of the respective goldfish proteins indicate that both goldfish TNFα1 and TNFα2 bind either TNF-R1 or TNF-R2 (199). Notably, recombinant sea bream TNFα (191), and the goldfish recombinant TNFα1, TNFα2, TNF-R1, and TNF-R2 all adopt homo-dimeric conformations and associate as dimers as opposed to the trimeric confirmations seen in the mammalian TNF ligands and receptors (199). Similarly, the grass carp TNFα ligand and TNF-R1 also associate as dimers (201). Interestingly, dimerized forms of the mammalian TNF-R1 have been observed (202–204) while the mammalian TNF receptor superfamily member, neurotrophin receptor (p75/NTR), is structurally similar to the teleost TNF-R1 and binds to the NTR ligand as a dimer (205, 206).
By studying the TNF systems of teleost fish, we may garner greater insights into the evolutionary origins of these important and evolutionarily conserved cytokines and receptors. Indeed the importance of the teleost TNFα proteins to their immune defenses is underlined by the fact that a number of diverse viral fish pathogens encode decoy TNF receptors (207–210).
Macrophages and Acute Phase Proteins (APPs) of Bony Fish
During inflammation, activated macrophages secrete cytokines and oxidative radicals that modulate the production of APPs by hepatic cells [reviewed by Gruys et al. (211)]. These APPs opsonize pathogens, activate complement, neutralize enzymes, and scavenge free hemoglobin and radicals.
Acute phase proteins rapidly increase in the blood early after exposure to pathogens or during early inflammatory response. For example, blood levels of C-reactive protein (CRP) may increase as much as 1000-fold and 50% increases in complement proteins and ceruloplasmin (Cp) have been observed. The activation of hepatocytes also results in decreased levels of serum transferrin, cortisol-binding globulin, zinc, iron, albumin, and retinol, as well as reduction of free hormones in the blood (212).
While viral infections induce modest acute phase responses (213), bacterial infections elicit potent production of these soluble mediators (211, 214–216). Upon recognition of LPS, monocytes and macrophages also produce gratuitous amounts of pro-inflammatory cytokines (214, 216–219). The termination of APP production is controlled by pro-inflammatory cytokines secreted by macrophages (220, 221).
Bony fish have fully functional repertoires of APPs, which are shared with their mammalian counterparts, as well as additional APPs that are unique to teleosts. The serum-CRP levels of salmonids have been used as indicators of stress in response to xenobiotics (222–224), and protozoan infections (225). The infection of goldfish with Trypanosoma carassii increased expression of Cp, crp, and serum amyloid A (saa), in the liver, particularly during the early phases of the infection (first 14 days of infection) (225). Serum amyloid-A (SAA) and a serum amyloid P-CRP-like pentraxin proteins have also been identified in salmonids (226), and goldfish (227). Aeromonas salmonicida infection of salmon also induced increased levels of SAA protein (226), while goldfish recombinant SAA was shown to induce increased gene expression of il12p40 and il1b, and was chemotactic to primary goldfish macrophages (227). It has also been demonstrated that similar to mammals, trout CRP was capable of activating complement (228).
The salmon saa was shown to be upregulated in hepatocytes after their exposure to supernatants from LPS-activated macrophages, or recombinant TNFα, IL-1β, or IL-6 (229). Interestingly, while LPS stimulations increased the expression of the fish pentraxin, A. salmonicida infections downregulated the expression of this gene, suggesting that pentraxin may be a “negative” APP (226, 230).
A selective subtractive hybridization (SSH) study of hepatic transcripts in unchallenged and bacterially challenged trout confirmed that a fully functional, broad-repertoire acute phase response exists in teleosts (378). Furthermore, after exposure to distinct pathogens, trout produce overlapping but partially distinct profiles of APPs (231). Catfish also have a well-developed acute phase response following bacterial infections leading to a 50-fold increase in the expression of some of the genes that encode APPs (232). In zebrafish, SSH analysis revealed that zebrafish infected with A. salmonicida and Staphylococcus aureus possess overlapping as well as unique APPs to those reported in mammals (233).
Macrophages and Complement
During a pathogen insult or PAMPs-induced inflammatory responses, there is a significant increase in blood complement levels [reviewed by Mastellos et al. (234) and Markiewski and Lambris (235)]. Most of the mammalian complement components exist in bony fish [reviewed by Nonaka (236)]. When compared with mammals, birds, and amphibians; teleosts have a full set of complement genes with the exception of Factor D, and the absence of MASP-1 and MASP-2 (236). Thus bony fish have multiple forms of several complement components including C3 and C5 proteins (237–241).
Fish complement components have similar pro-inflammatory roles akin to those of mammals. The anphylatoxin, C5a, has chemo-attractive activity (237, 240) and trout C3a enhances fish leukocyte phagocytosis (238, 241). In addition trout C3a, C4a, and C5a has been shown to be chemo-attractive to head kidney phagocytes and PBLs, and enhance phagocytosis of kidney leukocytes (242). The teleost complement biology has been fully addressed in a review by Sunyer et al. (243).
Antimicrobial Roles of Teleost M1 Macrophages
Phagocytosis
Phagocytosis is the primordial defense mechanism of all metazoan organisms. During the inflammatory response monocytes/macrophages and neutrophils, undergo phagocytosis mediated via phagocytic receptors or hydrophobic interactions of the phagocyte membrane and the target particles. Once activated, phagocytes release numerous preformed or newly synthesized inflammatory mediators, and are equipped with an armamentarium of antimicrobial responses primarily focused on the pathogens enclosed in the phagolysosomes. Potent antimicrobial compounds generated by activated phagocytes include degradative enzymes (proteases, nucleases, phosphatases, and lipases) and antimicrobial peptides (basic proteins and neutrophilic peptides), which mediate the destruction of phagocytosed pathogens (244–249).
Respiratory Burst Response
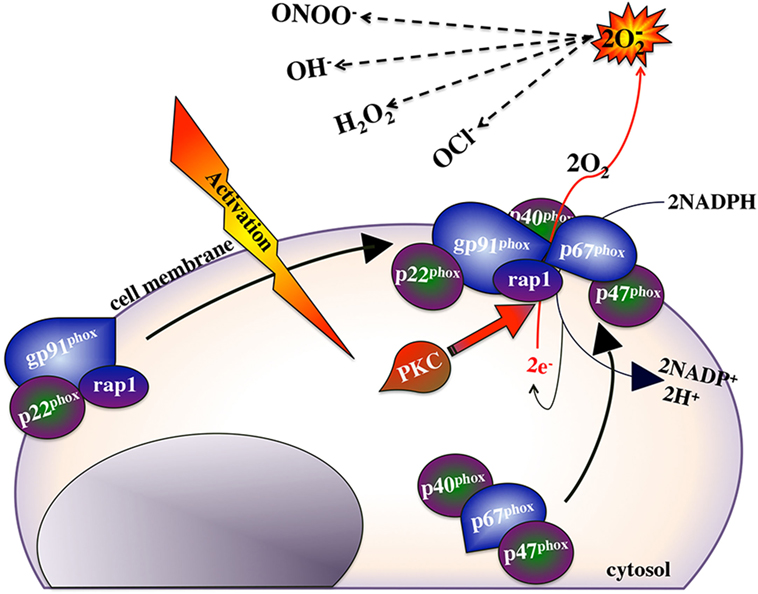
Macrophage ROS response is a hallmark of these cells’ antimicrobial armamentarium and the efficacy of this response often reflects on the ability of macrophages to destroy internalized microorganisms. This response culminates from the assembly of a multicomponent enzymatic complex, the nicotinamide adenine dinucleotide phosphate (NADPH, Figure 2) oxidase on the plasma and phagosome membranes, resulting in the transfer of electrons from NADPH to molecular oxygen and thus the production of a superoxide anion (250). In turn, the generated superoxide anions may be converted into other antimicrobial ROS such as hydrogen peroxide (H2O2), hydroxyl radical (OH⋅), and hypercholorus acid (251, 252). The NADPH oxidase complex has six interactive subunits including the cytosolic phagosome oxidases (p40phox, p47phox, and p67phox), and a guanosine triphosphatase Rac 1 or Rac 2, which are mobilized to the gp91phox and p22phox subunits that are located in the plasma membrane (253–258). All of these NADPH oxidase components have been identified in teleosts and fish macrophage ROS responses has been well documented in contexts of PAMP stimulation (259–262), antimicrobial responses (263–265), and recombinant cytokines stimulation such as with TNFα (183, 184, 266), IFNγ (136, 138, 148), and CSF-1 (32).
Tryptophan Degradation
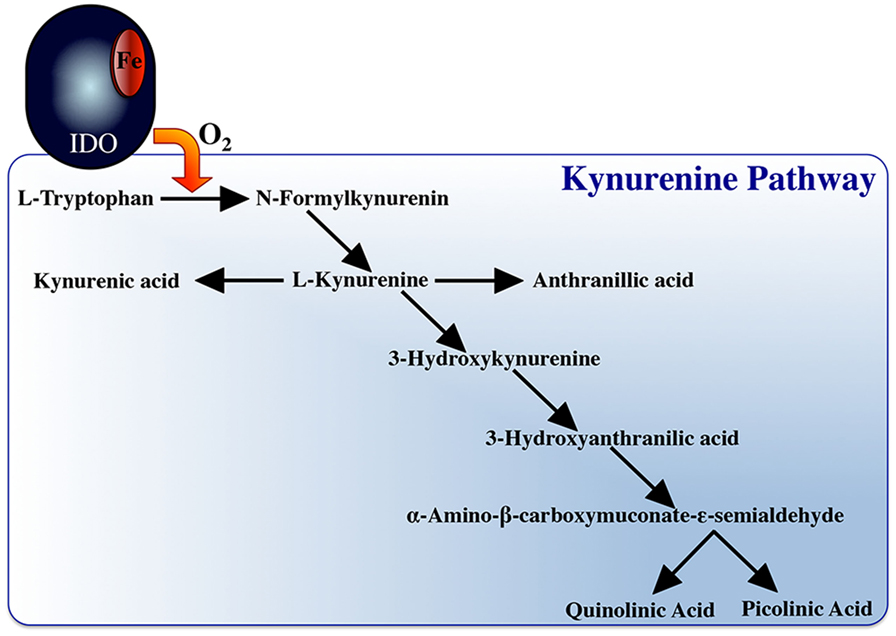
Another hallmark of M1 macrophages is their capacity to deplete local tryptophan levels through their upregulated expression of the indoleamine 2,3-dioxygenase (IDO) enzyme (267) (Figure 1), which catalyzes this process (268). IDO-mediated tryptophan degradation is closely linked to macrophage antimicrobial responses but also to their immunoregulatory functions, as this tryptophan degradation results in the production of metabolites such as kynurenins ((269), Figure 3), which may inhibit T cell proliferation. IFNγ-stimulation of macrophages has been closely linked to inducing the mammalian macrophage IDO response (270–273).
The teleost IDO orthologs (renamed proto-IDOs) are less effective at tryptophan degradation than the mammalian IDOs (274), bringing to question whether these fish enzymes have distinct substrates. Interestingly, Mycobacterium marinum-challenged goldfish macrophages upregulate their proto-ido gene expression (275), suggesting a possible M1 role for this fish enzyme.
Nitric Oxide Response
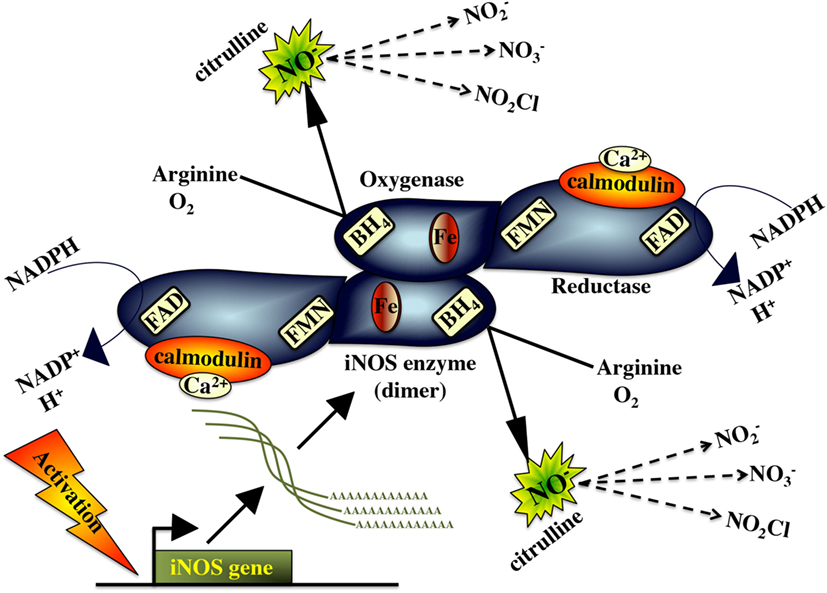
Classically activated M1 macrophages possess high levels of the inducible nitric oxide synthase enzyme (iNOS/NOS2), which catalyzes the conversion of L-arginine to L-citrulline, resulting in the production of nitric oxide (NO) (276) (Figure 4). As such, iNOS expression serves as a marker of M1 macrophage activation, which may be enhanced by macrophage stimulation with IFNγ, TNFα, and/or microbial compounds (e.g., LPS) (106). The parallel production of superoxide and NO can also result in the formation of peroxynitrite (ONOO-), which is a potent antiparasitic/antimicrobial agent (277). The immune mechanism governing the teleost macrophage inducible nitric oxide (NO) appears to be well conserved to those described in mammals.
Akin to its mammalian counterpart, the fish iNOS has putative binding sites for heme, calmodulin, flavine mononucleotide, flavine adenine dinucleotide tetrahydrobiopterin, and NADPH, indicating that this is a highly conserved enzyme (278). The fish macrophage iNOS gene is induced by antimicrobial and inflammatory stimuli such as PAMPs/pathogen recognition (10, 11, 278, 279), pro-inflammatory cytokines (138, 139, 183, 187) and cleaved transferrin products (280, 281). In turn, effective fish macrophage nitric oxide production is integral to fish antimicrobial immunity to a range of pathogens (282–287).
Sequential Induction of Macrophage Antimicrobial Responses
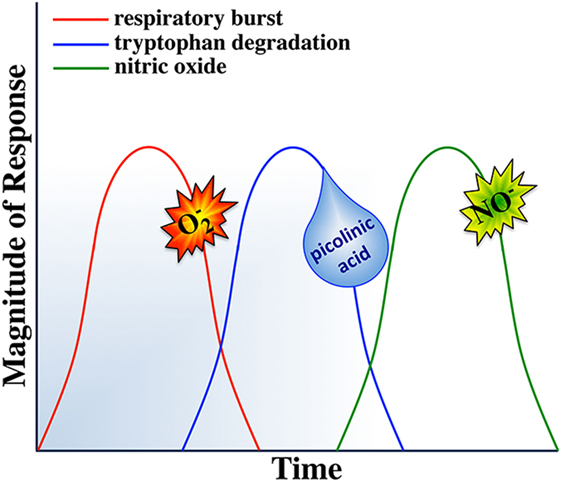
While mammalian macrophages are thought to be able to undergo simultaneous ROI and NO responses (288), there are several reports suggesting that teleost (primarily cyprinid) fish mount and sequentially deactivate their antimicrobial responses (138, 184, 260, 289, 290). We are aware of only one report describing sequential mammalian macrophage production of ROS followed by NO (291). However, the interdependence of the respective mammalian macrophage respiratory burst, tryptophan degradation, and nitric oxide responses suggest that sequential regulation of macrophage antimicrobial responses is not a strategy that is unique to teleosts and may be a predetermined fail-safe component of all vertebrate macrophage antimicrobial responses.
The respiratory burst and nitric oxide responses are thought of, as two independent macrophage microbicidal mechanisms, where in the induction of one does not depend on the induction of the other (288). However, both responses may be linked to tryptophan degradation. IDO activation requires reduction of its ferric (Fe3+) heme to ferrous (Fe2+) heme and there has been some contention regarding the source(s) of electrons used toward this reduction of the IDO heme (292, 293). Interestingly, a prevailing theory suggests that the superoxide anion, derived from the respiratory burst response, is in turn shunted into this enzymatic pathway, serving as this electron source (270, 273, 294, 295). It is interesting to consider that the sequential induction of the respiratory burst response before tryptophan degradation would ensure sufficient quantities of superoxide as a substrate for IDO activity and in turn would repurpose any remaining superoxide anions that had not reacted with the pathogen, thereby also minimizing bystander host cell damage. This notion is supported by the fact that the metabolites from tryptophan degradation are potent scavengers of ROS (296, 297). This in mind, simultaneous induction of macrophage tryptophan degradation and the respiratory burst response would thus be an overall inefficient microbicidal strategy, as the ROS would be actively scavenged by tryptophan catabolites. Thus, sequentially mounting these responses (Figure 5) would maximize the targeted effects of the respective responses.
Notably, macrophage tryptophan degradation appears to also be coupled to production of nitric oxide. Picolinic acid, a catabolite of tryptophan degradation (Figure 3), synergizes with IFNγ to induce nitric oxide production in murine macrophages (298–302). Picolinic acid exerts this nitric oxide inducing potential via a hypoxic responsive element located in the 5’ flanking region of the murine iNOS gene, while mutation or deletion of this promoter sequence impairs picolinic acid-induced gene transcription of iNOS without affecting induction of nitric oxide synthase by LPS (300). Thus, we propose that staggering the kinetics of macrophage tryptophan degradation and nitric oxide production would ensure sufficient quantities of picolinic acid toward the synergistic induction of nitric oxide. In turn, if the respiratory burst response was concomitantly induced with nitric oxide production, then picolinic acid could not exert its nitric oxide inducing effects, as the respiratory burst creates an extremely hyperoxic microenvironment. We thus suggest that the induction of tryptophan degradation before nitric oxide production would facilitate the establishment of a hypoxic microenvironment due to the tryptophan catabolites actively scavenging reactive oxygen intermediates, permitting picolinic acid to augment nitric oxide production by macrophages.
Nitric oxide appears to be the terminal microbicidal response of vertebrate macrophages. In addition to its potent killing effects, nitric oxide is a deactivator of specific enzymes involved in macrophage cytotoxic reactions. Interestingly, NO inhibits both protein kinase C (needed for initiating the ROI response; Figure 2) and IDO enzymes involved in the activation of the respiratory burst and tryptophan degradation, respectively (303, 304). Moreover, nitric oxide acts as a negative feedback inhibitor of its own synthesis (305, 306). Therefore, simultaneous induction of nitric oxide, respiratory burst and tryptophan degradation responses would antagonize PKC and thus NADPH oxidase activation (Figure 2) and the IDO enzyme. By sequentially inducing the nitric oxide response, subsequent to the respiratory burst and tryptophan degradation responses would ensure that each of these responses would be maximally induced and terminated in a timely manner, thus maximizing these respective antimicrobial responses and minimizing off-target effects of each response.
Based on the above and as outlined in Figure 5, we propose that such sequential induction and deactivation of macrophage antimicrobial responses may represent an important and presently poorly explored component of macrophage defenses. As activated macrophages are highly cytotoxic, the interdependence and temporal segregation of their individual microbicidal responses likely represents an inherent way to minimize host cell damage and concomitantly to maximize pathogen elimination. For example, pathogenic microorganisms that are susceptible to ROI are rapidly killed upon phagocytosis by activated macrophages while those pathogens that are resistant to oxidative burst, are often susceptible to subsequent nutrient deprivation and/or antimicrobial attacks. Indeed, ablating the macrophage respiratory burst response while shunting the produced superoxide anion into tryptophan degradation and the subsequent utilization of the picolinic acid from this response toward NO production (Figure 5) would maximize the effectiveness of each respective response. This would allow macrophages to divert and target their metabolic energy into distinct, targeted and timely antimicrobial assaults.
The proposed model shown in Figure 5 does not define macrophage activation in the context of a given individual macrophage, and indeed individual macrophages do not necessarily have to cycle through all of the above responses. Moreover, while much contention remains regarding the functionality of dipartite mammalian macrophage subsets, teleosts clearly possess macrophage sub-populations exhibiting dramatically different kinetics of activation and distinct antimicrobial capacities (9, 148, 184, 290). Notably, cyprinid kidney-derived monocyte-like cultures are considerably more proficient producers of ROS whereas the maturation of these cultures into predominantly macrophage-like cells coincides with their loss of respiratory burst capacities and a concomitant gain of significantly more robust NO responses (184). Presumably, sub-populations of macrophages with distinct antimicrobial potentials coordinate the sequential induction of macrophage antimicrobial responses in vivo.
It is unclear why despite considerably more rigorous investigation of the mammalian macrophage, there is more evidence of sequential macrophage antimicrobial responses in teleosts. The central M1/classical activation strategies of mammals and teleosts are best framed by their respective functional polarization by IFNγ. As described above, mammalian species possess single IFNγ molecules that are important for the activation of M1 macrophage ROI and NO responses (Figure 1). Intriguingly, many teleost fish possess multiple distinct IFNγ proteins, some of which appear to be potent elicitors of the macrophage ROS, but not NO responses whereas others elicit robust NO production but meager ROIs (148). Thus, we argue that these fish species may have evolved to generate multiple distinct M1 macrophage populations, here denoted as M1a and M1b (Figure 1). As an extension of this notion, we argued that fish may have evolved this relatively elaborate classical macrophage activation strategy in order to better coordinate, and when needed, segregate their respective macrophage antimicrobial responses.
Activation of Alternative/M2 Teleost Macrophages
Interleukin-4/13
M2 macrophages have ‘anti-inflammatory’, or ‘pro-healing’ phenotypes and the most extensively characterized M2-polarizing agents (sometimes called M2a) are the IL-4 and IL-13 cytokines (Figure 1), which are typically produced by Th2 cells, eosinophils, basophils, NK-T cells and certain macrophages subsets (307). IL-4 binds to the IL-4 receptor-alpha and either the IL-4 receptor-gamma or the IL-13 receptor-alpha1 chains, culminating in Jak1, Jak3, and Stat6 downstream signaling (104). IL-13 also ligates the IL-13 receptor-alpha2 chain (104). Either of these M2 stimuli result in increased of expression/production of a number of hallmark M2 macrophage components including transglutaminase 2, prostaglandin-endoperoxide synthase, transcription factors IRF4, macrophage mannose receptor, and suppressor of cytokine signaling 1 (SOCS1), all of which are present in fish but await to be functionally linked to teleost M2 macrophages (308–313).
Teleost possess IL-4/13A and IL-4/13B genes with sequence homology to both the mammalian IL-4 and IL-13 cytokines (314). These fish cytokines are thought to have arisen from genome/gene duplication events, and are present in distinct copies in different fish species (315). Paralogs of IL-4Rα, IL-13Rα1 and IL-13Rα2 have also been identified in teleosts (316, 317), while the recombinant fish IL-4/13A induces B and T cell expansion in an IL-13Rα-dependent manner (318, 319), suggesting that the roles of these fish cytokines possess the immune roles of their mammalian counterparts. The fish IL-4/13A and IL-4/13B are thought to play the M2/anti-inflammatory roles attributed to the mammalian IL-4 and IL-13 (320) and the trout, seabass, grass carp and goldfish recombinant IL-4/13A and IL-4/13B possess many of these anti-inflammatory roles including the upregulation of immunosuppressive genes (TGF-β, IL-10, SAP1, and SOCS3); dampening of pro-inflammatory cytokine gene expression (TNFα, IL-1β, and IFNγ); as well as elevating macrophage/kidney phagocyte arginase gene expression and arginase activity (321–324). Notably, a true Th2 locus has been identified in spotted gar, consisting of RAD50, IL-4/13 and IL-3/IL-5/GM-CSF (IL-5) (325) while the constitutively high expression of trout and salmon IL4/13A in the thymus, skin and gill tissues have been attributed to immunological tolerance and thus a Th2-like response (320).
Arginase
The enhanced capacity to metabolize L-arginine marks an important paradigm between M1 and M2 macrophages and underlines the M2 macrophage. This is intuitive, as M1 macrophage armamentarium is known for its elevated iNOS enzyme, which converts L-arginine to L-citrulline and NO. By contrast, the M2 macrophage arginase enzyme converts L-arginine to L-ornithine and urea (326, 327). The tissue repair capacities of these M2 macrophages in turn reflect their production of L-ornithine, which serves as a precursor for polyamines and proline components of collagen, during tissue repair (328). Notably, the products of these iNOS and arginase enzymatic pathways serve as reciprocal inhibitors of these antagonistic enzymes, promoting the respective M2 or M1 macrophage phenotypes (329).
Mammals possess two arginase isoforms, of which the macrophage gene expression of arginase-1 is induced by IL-4 and IL-13 (330). By contrast, macrophage arginase-2 gene expression is upregulated by IL-10 and LPS (331). Fish possess both arginase-1 and arginase-2 (332) and like mammals the fish M1/M2 paradigm is outlined by respectively elevated macrophage inos and arginase genes (10, 11, 279). By contrast to the mammalian M2 macrophages, carp alternative macrophage activation results in the induction of arginase-2 rather than arginase-1 expression (10). The facets of fish macrophage M2 polarization and the roles of arginase-2 to in this process have been thoroughly reviewed (190, 333).
GCs and Interleukin-10
Glucocorticoids and IL-10 stimulation of macrophages culminates in a unique regulatory macrophage phenotype, otherwise known as M2c. GCs diffuse across plasma membranes, resulting in alterations to the expression of a plethora of immune-related genes, which results in these M2c macrophage transcriptional profiles that are distinct from those seen in IL-4/IL-13-stimulated macrophages (334, 335). These M2c macrophage transcriptional changes include decreased inflammatory cytokine gene expression and dampening of ROS production. In line with the immunosuppressive nature of GCs, cortisol increases fish susceptibility to diseases (335, 336) and inhibits fish macrophage NO production (337). Moreover, the simultaneously of fish macrophage cell lines with combined pro-inflammatory stimuli and cortisol results in elevated il10 gene expression (13), indicating that the cortisol treatment overrides the inflammatory stimuli.
The mammalian IL-10 cytokine signals through a receptor complex composed of IL-10 receptors 1 (IL-10R1) and 2 (IL-10R2), leading to downstream STAT3 activation, which results in decreased gene expression of pro-inflammatory cytokines (338). Macrophage IL-10 production may be elicited by TLR agonists, GCs, and C-type lectins (307). Fish IL-10R1 has been identified in several cyprinids (339, 340), while the IL-10R2 has been reported in salmonids (341). Consistent with the mammalian counterpart, the goldfish recombinant IL-10 down-regulates macrophage ROS responses and inflammatory gene expression (275).
The Macrophage Bridge Between the Innate and Adaptive Immunity
In addition to their roles in early antimicrobial responses, macrophage-lineage cells are crucial to bridging the innate and adaptive arms of the vertebrate immune response. To this end, mammalian macrophages present intracellular pathogen-derived antigens to conventional CD8+ cytotoxic T cells via the MHC I pathway (342); extracellular antigens to CD8+ T cells in the context of MHCI by means of antigen cross-presentation (343) and extracellular antigens to conventional CD4+ T helper cells by means of MHCII complex (344). In addition, myeloid cells may present non-protein antigens to unconventional lymphocytes, such as lipid antigens in the context of non-classical MHCI (CD1) to invariant T cells and NK-T cells (345). Moreover, macrophages readily clean up antibody-opsonized pathogens through Fc-receptor-mediated phagocytosis (346). The molecular mechanisms by which teleost fish macrophages bridge the innate and adaptive arms of their respective immune responses are by far the most poorly understood.
Teleost Antigen Presentation
The fish (salmonid) MHCI peptide-loading complex appears to be fundamentally and functionally similar to that of mammals and the macrophage-like (RTS11) trout cell line has been demonstrated to assemble this antigen presentation complex (347–349). Moreover, trout appear to possess an alternatively spliced variant of MHCI loading glycoprotein, tapasin (349), which is believed to serve as additional regulatory mechanisms in the fish MHCI antigen presentation pathway. While some fish species such as medaka, sharks (350) and zebrafish (351) possess considerable polymorphism within their respective MHCI loci, other species such as Atlantic salmon do not have significant polymorphisms within their classical MHC I antigen processing genes (352). Interestingly, some of the other salmon MHC I assembly and antigen processing genes have been retained as functional duplicates (352). It is thought that these duplicated gene originated from the second vertebrate genome duplication event and are now providing various fish (and some tetrapods such as frogs and birds) with the potential of several different peptide-loading complexes (352).
Several teleost lineages have independently lost key components associated with mammalian antigen presentation and immunological memory including MHCII and CD4 (353–355), although these species exhibit effective immune responses, suggesting that they have evolved alternative immunological strategies for dealing with repeat infections. Moreover, recent genome assembly efforts concomitant with expression analyses have yielded the reconstruction of the evolutionary history of the MHCI (356) and MHCII (357) gene families, demonstrating that teleosts MHC loci have undergone a complex series of gene and genome duplications, culminating in extensive variation in MHC structure and diversity across these animals (358). These distinct teleost species have undoubtedly evolved distinct antigen presentation strategies coinciding with their great diversity across MHCI and II loci. Little is presently know regarding the roles of professional antigen presenting cells such as macrophages in these respective species and it will be most interesting to learn how such cells are integrated within these diverse immune systems.
Distinct fish species also possess several disparate lineages of non-classical MHCs (358), the linkage of which is now believed to have separated before the emergence of tetrapods (359). However, the roles of teleost macrophages and other professional antigen presenting cells in presenting novel antigens in the context of these molecules remain to be explored.
Teleost DCs
Myeloid-lineage DCs represent heterogeneous populations of professional antigen presenting cells that share a common myeloid progenitor (macrophage-dendritic cell progenitor) with macrophages and are integral to linking the innate and adaptive immune responses (360). Teleosts appear to possess functional analogs to the mammalian DCs and in particular, salmonids have been documented to possessing putative DCs. For example, salmon possess DC-like cells that express MHCII and CD83 (DC marker), are highly phagocytic and exhibit characteristic DC morphology (361). Trout also clearly possess DC-like cells expressing MHCII and other antigen presentation components, many DC markers (362) and exhibiting robust antigen presentation and lymphocyte activation capacities (363). Moreover, trout appear to possess DCs with cross-presentation capacities that express the same hallmark markers seen on the mammalian DCs specialized to antigen cross-presentation (364, 365). Similarly, the cyprinid zebrafish have been shown to possess cells expressing hallmark DC markers and displaying the capacity to present antigens and induce the proliferation of fish CD4+ T cells (366).
The Link Between Teleost Innate and Antibody Responses
It is presently not clear what roles teleost antibodies play in the opsonization of pathogens that enhance macrophage phagocytosis and the canonical Fc receptors responsible for this process in mammals have not been fully elucidated in teleosts (367). However, there are at least five distinct immunoglobulin domain-containing multi-gene receptor families with some structural and signaling motifs seen in the mammalian Fc receptors (368). Moreover, as members of at least one of this family (LITRs) appear to play roles in phagocytosis (369, 370), it is conceivable that members of this, as well as the other receptor families may function as fish phagocytic receptors for antibody-opsonized targets.
While teleost orthologs to the mammalian Fc receptors remain elusive, teleosts are now known to encode poly Ig receptors (pIgRs) that are capable of binding to fish antibodies (371, 372) and appear to be involved in phagocytosis (373) but are not expressed on fish macrophages or B cells (371). It will be interesting to learn whether distinct subsets of fish phagocytes may acquire the expression of pIgRs immune stimuli.
It is notable that cartilaginous fish (sharks) possess IgM-mediated opsonization and cytotoxicity, which is mediated by granulocytes rather than macrophages (374). Turbot macrophage phagocytosis of yeast and beads was greatly enhanced by opsonization with turbot Ig-containing serum fraction however, Ig-opsonized microsporidian spores were not taken up at a greater rate than non-opsonized spores (375). Similarly, brook trout macrophages phagocytosis of A. salmonicida was not enhanced when following opsonization of the bacteria by specific fish antibodies although complement-mediated opsonization significantly enhanced bacterial uptake (376). It will be interesting to learn whether the teleost macrophage apparent lack of hallmark Fc receptors reflects in the above observations or whether bony fish macrophages are capable of undergoing antibody-mediated phagocytosis under distinct conditions and through distinct molecular mechanisms.
Concluding Remarks
Akin to the vast heterogeneity of functionally desperate macrophage subsets observed across mammals, teleost fish appear to possess both a spectrum of functionally distinct macrophage subsets as well as a plethora of potential molecular drivers of these distinct lineages. Moreover and in consideration of the strikingly distinct teleost physiologies, evolutionary and pathogenic pressures as well different repertoires of candidate macrophage differentiation factors, these organisms may well utilize (at least partially) distinct macrophage differentiation and activation strategies. It is notable that while many fish species possess multiple isoforms of key macrophage cytokines, functional studies of these moieties have often been limited to one of the several isoforms and have addressed similarities to the mammalian counterparts whilst overlooking some potential functional differences. Indeed, distinct whole genome duplication events and the ploidy of respective fish species can be seen in disparate cytokine copy-number repertoires amongst even closely related fish species (377). These differences are exemplified in copy numbers of hallmark macrophage cytokines such as IFNγ and TNFα across distinct fish. It is generally assumed that the roles of these respective molecules are conserved to those of mammals. However, it is likely that the retention of multiple isoforms within a particular fish species and the often seen expression differences between these fish cytokine isoforms indicate non-overlapping and possibly novel roles for these respective immune mediators. A greater understanding of the mechanisms of fish macrophage antimicrobial immunity is warranted toward aquacultural applications and for the sake of fundamental research. With greater availability of both fish-specific reagents and genomic resources, the time is ripe for advancing our understanding of these processes.
Author Contributions
LG, BK, AY, JH, JX, and MB participated in writing the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by the Natural Sciences and Engineering Council of Canada (NSERC; Grant No. RGPIN-2014-06395) to MB. LG thanks the George Washington University for financial support in the form of laboratory start-up funds. BK thanks George Washington University for teaching assistantship support. JH was supported by an NSERC PGS-D doctoral scholarship and JX by China Scholarship Council studentship and Alberta Innovates doctoral scholarship.
References
1. Amici SA, Dong J, Guerau-De-Arellano M. Molecular mechanisms modulating the phenotype of macrophages and microglia. Front Immunol (2017) 8:1520. doi:10.3389/fimmu.2017.01520
2. Vergadi E, Ieronymaki E, Lyroni K, Vaporidi K, Tsatsanis C. Akt signaling pathway in macrophage activation and M1/M2 polarization. J Immunol (2017) 198:1006–14. doi:10.4049/jimmunol.1601515
3. Hodgkinson JW, Grayfer L, Belosevic M. Biology of bony fish macrophages. Biology (Basel) (2015) 4:881–906. doi:10.3390/biology4040881
4. Zhou D, Huang C, Lin Z, Zhan S, Kong L, Fang C, et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal (2013) 26:192–7. doi:10.1016/j.cellsig.2013.11.004
5. Rieger AM, Hall BE, Barreda DR. Macrophage activation differentially modulates particle binding, phagocytosis and downstream antimicrobial mechanisms. Dev Comp Immunol (2010) 34:1144–59. doi:10.1016/j.dci.2010.06.006
6. Barreda DR, Neumann NF, Belosevic M. Flow cytometric analysis of PKH26-labeled goldfish kidney-derived macrophages. Dev Comp Immunol (2000) 24:395–406. doi:10.1016/S0145-305X(99)00059-2
7. Neumann NF, Stafford JL, Belosevic M. Biochemical and functional characterisation of macrophage stimulating factors secreted by mitogen-induced goldfish kidney leucocytes. Fish Shellfish Immunol (2000) 10:167–86. doi:10.1006/fsim.1999.0236
8. Mills CD, Ley K. M1 and M2 macrophages: the chicken and the egg of immunity. J Innate Immun (2014) 6:716–26. doi:10.1159/000364945
9. Grayfer L, Hodgkinson JW, Belosevic M. Antimicrobial responses of teleost phagocytes and innate immune evasion strategies of intracellular bacteria. Dev Comp Immunol (2014) 43:223–42. doi:10.1016/j.dci.2013.08.003
10. Joerink M, Savelkoul HFJ, Wiegertjes GF. Evolutionary conservation of alternative activation of macrophages: structural and functional characterization of arginase 1 and 2 in carp (Cyprinus carpio L.). Mol Immunol (2006) 43:1116–28. doi:10.1016/j.molimm.2005.07.022
11. Joerink M, Forlenza M, Ribeiro CM, De Vries BJ, Savelkoul HF, Wiegertjes GF. Differential macrophage polarisation during parasitic infections in common carp (Cyprinus carpio L.). Fish Shellfish Immunol (2006) 21:561–71. doi:10.1016/j.fsi.2006.03.006
12. Grayfer L, Hodgkinson JW, Hitchen SJ, Belosevic M. Characterization and functional analysis of goldfish (Carassius auratus L.) interleukin-10. Mol Immunol (2010) 48:563–71. doi:10.1016/j.molimm.2010.10.013
13. Castro R, Zou J, Secombes CJ, Martin SA. Cortisol modulates the induction of inflammatory gene expression in a rainbow trout macrophage cell line. Fish Shellfish Immunol (2011) 30:215–23. doi:10.1016/j.fsi.2010.10.010
14. Glasauer SMK, Neuhauss SCF. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics (2014) 289:1045–60. doi:10.1007/s00438-014-0889-2
15. Van Furth R, Cohn ZA, Hirsch JG, Humphrey JH, Spector WG, Langevoort HL. [Mononuclear phagocytic system: new classification of macrophages, monocytes and of their cell line]. Bull World Health Organ (1972) 47:651–8.
16. Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity (2013) 38:792–804. doi:10.1016/j.immuni.2013.04.004
17. Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol (2014) 14:392–404. doi:10.1038/nri3671
18. Yona S, Kim K-W, Wolf Y, Mildner A, Varol D, Breker M, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity (2013) 38:79–91. doi:10.1016/j.immuni.2012.12.001
19. Barreda DR, Hanington PC, Belosevic M. Regulation of myeloid development and function by colony stimulating factors. Dev Comp Immunol (2004) 28:509–54. doi:10.1016/j.dci.2003.09.010
20. Soza-Ried C, Hess I, Netuschil N, Schorpp M, Boehm T. Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc Natl Acad Sci U S A (2010) 107:17304–8. doi:10.1073/pnas.1004640107
21. Guilbert LJ, Stanley ER. Specific interaction of murine colony-stimulating factor with mononuclear phagocytic cells. J Cell Biol (1980) 85:153–9. doi:10.1083/jcb.85.1.153
22. Lichanska AM, Browne CM, Henkel GW, Murphy KM, Ostrowski MC, Mckercher SR, et al. Differentiation of the mononuclear phagocyte system during mouse embryogenesis: the role of transcription factor PU.1. Blood (1999) 94:127–38.
23. Dai X-M, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, et al. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood (2002) 99:111–20. doi:10.1182/blood.V99.1.111
24. Hanington PC, Wang T, Secombes CJ, Belosevic M. Growth factors of lower vertebrates: characterization of goldfish (Carassius auratus L.) macrophage colony stimulating factor-1. J Biol Chem (2007) 282:31865–72. doi:10.1074/jbc.M706278200
25. Wang T, Hanington PC, Belosevic M, Secombes CJ. Two macrophage colony-stimulating factor genes exist in fish that differ in gene organization and are differentially expressed. J Immunol (2008) 181:3310–22. doi:10.4049/jimmunol.181.5.3310
26. Grayfer L, Robert J. Distinct functional roles of amphibian (Xenopus laevis) colony-stimulating factor-1- and interleukin-34-derived macrophages. J Leukoc Biol (2015) 98:641–9. doi:10.1189/jlb.4AB0315-117RR
27. Chen X, Liu H, Focia PJ, Shim AH, He X. Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases. Proc Natl Acad Sci U S A (2008) 105:18267–72. doi:10.1073/pnas.0807762105
28. Hamilton TA, Zhao C, Pavicic PG Jr, Datta S. Myeloid colony-stimulating factors as regulators of macrophage polarization. Front Immunol (2014) 5:554. doi:10.3389/fimmu.2014.00554
29. Manos MM. Expression and processing of a recombinant human macrophage colony-stimulating factor in mouse cells. Mol Cell Biol (1988) 8:5035–9. doi:10.1128/MCB.8.11.5035
30. Rettenmier CW, Roussel MF. Differential processing of colony-stimulating factor 1 precursors encoded by two human cDNAs. Mol Cell Biol (1988) 8:5026–34. doi:10.1128/MCB.8.11.5026
31. Hanington PC, Hitchen SJ, Beamish LA, Belosevic M. Macrophage colony stimulating factor (CSF-1) is a central growth factor of goldfish macrophages. Fish Shellfish Immunol (2009) 26:1–9. doi:10.1016/j.fsi.2008.09.020
32. Grayfer L, Hanington PC, Belosevic M. Macrophage colony-stimulating factor (CSF-1) induces pro-inflammatory gene expression and enhances antimicrobial responses of goldfish (Carassius auratus L.) macrophages. Fish Shellfish Immunol (2009) 26:406–13. doi:10.1016/j.fsi.2008.12.001
33. Rieger AM, Hanington PC, Belosevic M, Barreda DR. Control of CSF-1 induced inflammation in teleost fish by a soluble form of the CSF-1 receptor. Fish Shellfish Immunol (2014) 41:45–51. doi:10.1016/j.fsi.2014.03.035
34. Rieger AM, Konowalchuk JD, Havixbeck JJ, Robbins JS, Smith MK, Lund JM, et al. A soluble form of the CSF-1 receptor contributes to the inhibition of inflammation in a teleost fish. Dev Comp Immunol (2014) 39:438–46. doi:10.1016/j.dci.2012.12.001
35. Williams H, Brenner S, Venkatesh B. Identification and analysis of additional copies of the platelet-derived growth factor receptor and colony stimulating factor 1 receptor genes in fugu. Gene (2002) 295:255–64. doi:10.1016/S0378-1119(02)00736-9
36. Chihara T, Suzu S, Hassan R, Chutiwitoonchai N, Hiyoshi M, Motoyoshi K, et al. IL-34 and M-CSF share the receptor Fms but are not identical in biological activity and signal activation. Cell Death Differ (2010) 17:1917–27. doi:10.1038/cdd.2010.60
37. Ma X, Lin WY, Chen Y, Stawicki S, Mukhyala K, Wu Y, et al. Structural basis for the dual recognition of helical cytokines IL-34 and CSF-1 by CSF-1R. Structure (2012) 20:676–87. doi:10.1016/j.str.2012.02.010
38. Liu H, Leo C, Chen X, Wong BR, Williams LT, Lin H, et al. The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1. Biochim Biophys Acta (2014) 1824:938–45. doi:10.1016/j.bbapap.2012.04.012
39. Baud’huin M, Renault R, Charrier C, Riet A, Moreau A, Brion R, et al. Interleukin-34 is expressed by giant cell tumours of bone and plays a key role in RANKL-induced osteoclastogenesis. J Pathol (2010) 221:77–86. doi:10.1002/path.2684
40. Chen Z, Buki K, Vaaraniemi J, Gu G, Vaananen HK. The critical role of IL-34 in osteoclastogenesis. PLoS One (2011) 6:e18689. doi:10.1371/journal.pone.0018689
41. Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, et al. Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity (2012) 37:1050–60. doi:10.1016/j.immuni.2012.11.001
42. Wang Y, Colonna M. Interkeukin-34, a cytokine crucial for the differentiation and maintenance of tissue resident macrophages and Langerhans cells. Eur J Immunol (2014) 44(6):1575–81. doi:10.1002/eji.201344365
43. Yamane F, Nishikawa Y, Matsui K, Asakura M, Iwasaki E, Watanabe K, et al. CSF-1 receptor-mediated differentiation of a new type of monocytic cell with B cell-stimulating activity: its selective dependence on IL-34. J Leukoc Biol (2014) 95:19–31. doi:10.1189/jlb.0613311
44. Grayfer L, Robert J. Divergent antiviral roles of amphibian (Xenopus laevis) macrophages elicited by colony-stimulating factor-1 and interleukin-34. J Leukoc Biol (2014) 96:1143–53. doi:10.1189/jlb.4A0614-295R
45. Yaparla A, Popovic M, Grayfer L. Differentiation-dependent antiviral capacities of amphibian (Xenopus laevis) macrophages. J Biol Chem (2018) 293(5):1736–44. doi:10.1074/jbc.M117.794065
46. Mo ZQ, Li YW, Zhou L, Li AX, Luo XC, Dan XM. Grouper (Epinephelus coioides) IL-34/MCSF2 and MCSFR1/MCSFR2 were involved in mononuclear phagocytes activation against Cryptocaryon irritans infection. Fish Shellfish Immunol (2015) 43:142–9. doi:10.1016/j.fsi.2014.12.023
47. Wang T, Kono T, Monte MM, Kuse H, Costa MM, Korenaga H, et al. Identification of IL-34 in teleost fish: differential expression of rainbow trout IL-34, MCSF1 and MCSF2, ligands of the MCSF receptor. Mol Immunol (2014) 53:398–409. doi:10.1016/j.molimm.2012.09.008
48. Barreda DR, Hanington PC, Stafford JL, Belosevic M. A novel soluble form of the CSF-1 receptor inhibits proliferation of self-renewing macrophages of goldfish (Carassius auratus L.). Dev Comp Immunol (2005) 29:879–94. doi:10.1016/j.dci.2005.02.006
49. Rieger AM, Havixbeck JJ, Belosevic M, Barreda DR. Teleost soluble CSF-1R modulates cytokine profiles at an inflammatory site, and inhibits neutrophil chemotaxis, phagocytosis, and bacterial killing. Dev Comp Immunol (2015) 49:259–66. doi:10.1016/j.dci.2014.12.003
50. Janeway CA Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol (1989) 54(Pt 1):1–13. doi:10.1101/SQB.1989.054.01.003
51. Hansen JD, Vojtech LN, Laing KJ. Sensing disease and danger: a survey of vertebrate PRRs and their origins. Dev Comp Immunol (2011) 35(9):886–97. doi:10.1016/j.dci.2011.01.008
52. Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature (2002) 418:191–5. doi:10.1038/nature00858
53. Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol (2008) 8:279–89. doi:10.1038/nri2215
54. Yamasaki S, Ishikawa E, Sakuma M, Hara H, Ogata K, Saito T. Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat Immunol (2008) 9:1179–88. doi:10.1038/ni.1651
55. Parker LC, Prince LR, Sabroe I. Translational mini-review series on toll-like receptors: networks regulated by toll-like receptors mediate innate and adaptive immunity. Clin Exp Immunol (2007) 147:199–207. doi:10.1111/j.1365-2249.2006.03203.x
56. Beutler B, Rehli M. Evolution of the TIR, tolls and TLRs: functional inferences from computational biology. Curr Top Microbiol Immunol (2002) 270:1–21.
57. Valanne S, Wang JH, Ramet M. The Drosophila toll signaling pathway. J Immunol (2011) 186:649–56. doi:10.4049/jimmunol.1002302
58. Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell (1996) 86:973–83. doi:10.1016/S0092-8674(00)80172-5
59. Keestra AM, De Zoete MR, Van Aubel RA, Van Putten JP. The central leucine-rich repeat region of chicken TLR16 dictates unique ligand specificity and species-specific interaction with TLR2. J Immunol (2007) 178:7110–9. doi:10.4049/jimmunol.178.11.7110
60. Temperley ND, Berlin S, Paton IR, Griffin DK, Burt DW. Evolution of the chicken toll-like receptor gene family: a story of gene gain and gene loss. BMC Genomics (2008) 9:62. doi:10.1186/1471-2164-9-62
61. Ishii A, Kawasaki M, Matsumoto M, Tochinai S, Seya T. Phylogenetic and expression analysis of amphibian Xenopus toll-like receptors. Immunogenetics (2007) 59:281–93. doi:10.1007/s00251-007-0193-y
62. Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, et al. The evolution of vertebrate toll-like receptors. Proc Natl Acad Sci U S A (2005) 102:9577–82. doi:10.1073/pnas.0502272102
63. Huang S, Yuan S, Guo L, Yu Y, Li J, Wu T, et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res (2008) 18:1112–26. doi:10.1101/gr.069674.107
64. Oshiumi H, Matsuo A, Matsumoto M, Seya T. Pan-vertebrate toll-like receptors during evolution. Curr Genomics (2008) 9:488–93. doi:10.2174/138920208786241234
65. Sepulcre MP, Alcaraz-Perez F, Lopez-Munoz A, Roca FJ, Meseguer J, Cayuela ML, et al. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J Immunol (2009) 182:1836–45. doi:10.4049/jimmunol.0801755
66. Sullivan C, Charette J, Catchen J, Lage CR, Giasson G, Postlethwait JH, et al. The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J Immunol (2009) 183:5896–908. doi:10.4049/jimmunol.0803285
67. Palti Y. Toll-like receptors in bony fish: from genomics to function. Dev Comp Immunol (2011) 35:1263–72. doi:10.1016/j.dci.2011.03.006
68. Pietretti D, Wiegertjes GF. Ligand specificities of toll-like receptors in fish: indications from infection studies. Dev Comp Immunol (2014) 43:205–22. doi:10.1016/j.dci.2013.08.010
69. Rebl A, Goldammer T, Seyfert H-M. Toll-like receptor signalling in bony fish. Vet Immunol Immunopathol (2010) 134:139–50. doi:10.1016/j.vetimm.2009.09.021
70. Li Y, Li Y, Cao X, Jin X, Jin T. Pattern recgnition receptors in zebrafish provide functional and evolutionary insight into innate immune signalling pathways. Cell Mol Immunol (2017) 14:80–9. doi:10.1038/cmi.2016.50
71. Zou J, Chang M, Nie P, Secombes CJ. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol Biol (2009) 9:85. doi:10.1186/1471-2148-9-85
72. Barber MR, Aldridge JR Jr, Webster RG, Magor KE. Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci U S A (2010) 107:5913–8. doi:10.1073/pnas.1001755107
73. Biacchesi S, Leberre M, Lamoureux A, Louise Y, Lauret E, Boudinot P, et al. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J Virol (2009) 83:7815–27. doi:10.1128/JVI.00404-09
74. Su J, Huang T, Dong J, Heng J, Zhang R, Peng L. Molecular cloning and immune responsive expression of MDA5 gene, a pivotal member of the RLR gene family from grass carp Ctenopharyngodon idella. Fish Shellfish Immunol (2010) 28:712–8. doi:10.1016/j.fsi.2010.01.009
75. Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP. Signaling in plant-microbe interactions. Science (1997) 276:726–33. doi:10.1126/science.276.5313.726
76. Cohn J, Sessa G, Martin GB. Innate immunity in plants. Curr Opin Immunol (2001) 13:55–62. doi:10.1016/S0952-7915(00)00182-5
77. Bertin J, Nir W-J, Fischer CM, Tayber OV, Errada PR, Grant JR, et al. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-κB. J Biol Chem (1999) 274:12955–8. doi:10.1074/jbc.274.19.12955
78. Inohara N, Koseki T, del Peso L, Hu YM, Yee C, Chen S, et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappa B. J Biol Chem (1999) 274:14560–7. doi:10.1074/jbc.274.21.14560
79. Ellis RE, Yuan JY, Horvitz HR. Mechanisms and functions of cell death. Annu Rev Cell Biol (1991) 7:663–98. doi:10.1146/annurev.cb.07.110191.003311
80. Peter ME, Heufelder AE, Hengartner MO. Advances in apoptosis research. Proc Natl Acad Sci U S A (1997) 94:12736–7. doi:10.1073/pnas.94.24.12736
81. Liu QA, Hengartner MO. The molecular mechanism of programmed cell death in C. elegans. Ann N Y Acad Sci (1999) 887:92–104. doi:10.1111/j.1749-6632.1999.tb07925.x
82. Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Núñez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J Biol Chem (2001) 276:4812–8. doi:10.1074/jbc.M008072200
83. Laing KJ, Purcell MK, Winton JR, Hansen JD. A genomic view of the NOD-like receptor family in teleost fish: identification of a novel NLR subfamily in zebrafish. BMC Evol Biol (2008) 8:42. doi:10.1186/1471-2148-8-42
84. Stein C, Caccamo M, Laird G, Leptin M. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol (2007) 8:R251. doi:10.1186/gb-2007-8-11-r251
85. Chen WQ, Xu QQ, Chang MX, Nie P, Peng KM. Molecular characterization and expression analysis of nuclear oligomerization domain proteins NOD1 and NOD2 in grass carp Ctenopharyngodon idella. Fish Shellfish Immunol (2010) 28:18–29. doi:10.1016/j.fsi.2009.09.012
86. Chang M, Wang T, Nie P, Zou J, Secombes CJ. Cloning of two rainbow trout nucleotide-binding oligomerization domain containing 2 (NOD2) splice variants and functional characterization of the NOD2 effector domains. Fish Shellfish Immunol (2011) 30:118–27. doi:10.1016/j.fsi.2010.09.014
87. Sha Z, Abernathy JW, Wang S, Li P, Kucuktas H, Liu H, et al. NOD-like subfamily of the nucleotide-binding domain and leucine-rich containining family receptors and their expression in channel catfish. Dev Comp Immunol (2009) 33:991–9. doi:10.1016/j.dci.2009.04.004
88. Rajendran K, Zhang J, Liu S, Kucuktas H, Wang X, Liu H, et al. Pathogen recognition receotprs in channel catfish: I. Identification, phylogeny and expression of NOD-like receptors. Dev Comp Immunol (2012) 37:77–86. doi:10.1016/j.dci.2012.02.004
89. Swain B, Basu M, Samanta M. Molecular cloning and characterization of nucleotide binding and oligomerization domain-1 (NOD1) receptor in the Indian Major Carp, rohu (Labeo rohita), and analysis of its inductive expression and down-stream signalling molecules following ligands exposure and Gram-negative bacterial infections. Fish Shellfish Immunol (2012) 32:899–908. doi:10.1016/j.fsi.2012.02.018
90. Swain B, Basu M, Sahoo BR, Maiti NK, Routray P, Eknath AE, et al. Molecular characterization of nucleotide binding and oligomerization domain (NOD)-2, analysis of its inductive expression and down-stream signaling following ligands exposure and bacterial infection in rohu (Labeo rohita). Dev Comp Immunol (2012) 36:93–103. doi:10.1016/j.dci.2011.06.018
91. Hou Q-H, Yi S-B, Ding X, Zhang H-X, Sun Y, Zhang Y, et al. Differential expression analysis of nuclear oligomerization domain proteins NOD1 and NOD2 in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol (2012) 33:1102–11. doi:10.1016/j.fsi.2012.08.015
92. Xie J, Hodgkinson JW, Katzenback BA, Kovacevic N, Belosevic M. Characterization of three NOD-like receptors of goldfish (Carassius auratus L.) and their role in antimicrobial responses of macrophages to Aeromonas salmonicida and Mycobacterium marinum. Dev Comp Immunol (2013) 39:180–7. doi:10.1016/j.dci.2012.11.005
93. Unajak S, Santos MD, Hikima J, Jung T-S, Kondo H, Hirono I, et al. Molecular characterization, expression and functional analysis of a nuclear oligomerization domain proteins subfamily C (NLRC) in Japanese flounder (Paralichthys olivaceus). Fish Shellfish Immunol (2011) 31:202–11. doi:10.1016/j.fsi.2011.05.007
94. Park SB, Hikima J, Suzuki Y, Ohtani M, Nho SW, Cha IS, et al. Molecular cloning and functional analysis of nucleotide-binding oligomerization domain 1 (NOD1) in olive flounder, Paralichthys olivaceus. Dev Comp Immunol (2012) 36:680–7. doi:10.1016/j.dci.2011.11.007
95. Li J, Gao Y, Xu T. Comparative genomic and evolution of vertebrate NOD1 and NOD2 genes and their immune response in miiuy croaker. Fish Shellfish Immunol (2015) 46:387–97. doi:10.1016/j.fsi.2015.06.026
96. Li S, Chen X, Hao G, Geng X, Zhan W, Sun J. Identification and characterization of a novel NOD-like receptor family CARD domain containing 3 gene in response to extracellular ATP stimulation and its role in regulating LPS-induced innate immune response in Japanese flounder (Paralichthys olivaceus) head kidney macrophages. Fish Shellfish Immunol (2016) 50:79–90. doi:10.1016/j.fsi.2016.01.029
97. Biswas G, Bilen S, Kono T, Sakai M, Hikima J. Inflammatory immune response by lipopolysaccharide-responsive nucleotide binding oligomerization domain (NOD)-like receptors in the Japanese pufferfish (Takifugu rubripes). Dev Comp Immunol (2016) 55:21–31. doi:10.1016/j.dci.2015.10.008
98. Angosto D, Lopez-Castejon G, Lopez-Munoz A, Sepulcre MP, Arizcun M, Meseguer J, et al. Evolution of inflammasome functions in vertebrates: Inflammasome and caspase-1 trigger fish macrophage cell death but are dispensable for the processing of IL-1beta. Inn Immun (2012) 18:815–24. doi:10.1177/1753425912441956
99. Angosto D, Mulero V. The zebrafish as a model to study the inflammasome. Inflammasome (2014) 1:2300X–2102X. doi:10.2478/infl-2014-0002
100. Xie J, Belosevic M. Functional characterization of receptro-interaciong serine/threonine kinase 2 (RIP2) of the goldfish (Carassius auratus L.). Dev Comp Immunol (2015) 48:76–85. doi:10.1016/j.dci.2014.09.006
101. Xie J, Belosevic M. Functional characterization of apoptosis-assiciated spec-like protein (ASC) of the goldfish (Carassius auratus L.) macrophages. Dev Comp Immunol (2016) 65:201–10. doi:10.1016/j.dci.2016.07.013
102. Xie J, Belosevic M. Characterization and functional assessment of the NLRC3-like molecule of the goldfish (Carassius auratus L.). Dev Comp Immunol (2017) 79:1–10. doi:10.1016/j.dci.2017.09.021
103. Gordon S. Alternative activation of macrophages. Nat Rev Immunol (2003) 3:23–35. doi:10.1038/nri978
104. Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity (2010) 32:593–604. doi:10.1016/j.immuni.2010.05.007
105. Robertsen B. The interferon system of teleost fish. Fish Shellfish Immunol (2006) 20:172–91. doi:10.1016/j.fsi.2005.01.010
106. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol (2008) 8:958–69. doi:10.1038/nri2448
107. O’Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity (2008) 28:477–87. doi:10.1016/j.immuni.2008.03.002
108. Zhou L, Cao X, Fang J, Li Y, Fan M. Macrophages polarization is mediated by the combination of PRR ligands and distinct inflammatory cytokines. Int J Clin Exp Pathol (2015) 8:10964–74. doi:10.1936-2625/IJCEP0011990
109. Van Der Vaart M, Spaink HP, Meijer AH, Van Der Vaart M, Spaink HP, Meijer AH. Pathogen recognition and activation of the innate immune response in zebrafish. Adv Hematol (2012) 2012(2012):e159807.
110. Wheelock EF. Interferon-like virus-inhibitor induced in human leukocytes by phytohemagglutinin. Science (1965) 149:310–1. doi:10.1126/science.149.3681.310
111. Belosevic M, Davis CE, Meltzer MS, Nacy CA. Regulation of activated macrophage antimicrobial activities. Identification of lymphokines that cooperate with IFN-gamma for induction of resistance to infection. J Immunol (1988) 141:890–6.
112. Staeheli P. Interferon-induced proteins and the antiviral state. Adv Virus Res (1990) 38:147–200. doi:10.1016/S0065-3527(08)60862-3
113. Stevenson MM, Tam MF, Belosevic M, Van Der Meide PH, Podoba JE. Role of endogenous gamma interferon in host response to infection with blood-stage Plasmodium chabaudi AS. Infect Immun (1990) 58:3225–32.
114. Kerr IM, Stark GR. The antiviral effects of the interferons and their inhibition. J Interferon Res (1992) 12:237–40. doi:10.1089/jir.1992.12.237
115. Krishnan L, Guilbert LJ, Wegmann TG, Belosevic M, Mosmann TR. T helper 1 response against Leishmania major in pregnant C57BL/6 mice increases implantation failure and fetal resorptions. Correlation with increased IFN-gamma and TNF and reduced IL-10 production by placental cells. J Immunol (1996) 156:653–62.
116. Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, et al. Immune response in mice that lack the interferon-γ receptor. Science (1993) 259(5102):1742–5. doi:10.1126/science.8456301
117. Wang ZE, Reiner SL, Zheng S, Dalton DK, Locksley RM. CD4+ effector cells default to the Th2 pathway in interferon gamma-deficient mice infected with Leishmania major. J Exp Med (1994) 179:1367–71. doi:10.1084/jem.179.4.1367
118. Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme IM. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med (1993) 178:2243–7. doi:10.1084/jem.178.6.2243
119. Fertsch D, Vogel SN. Recombinant interferons increase macrophage Fc receptor capacity. J Immunol (1984) 132:2436–9.
120. Berton G, Zeni L, Cassatella MA, Rossi F. Gamma interferon is able to enhance the oxidative metabolism of human neutrophils. Biochem Biophys Res Commun (1986) 138:1276–82. doi:10.1016/S0006-291X(86)80421-1
121. Cassatella MA, Bazzoni F, Flynn RM, Dusi S, Trinchieri G, Rossi F. Molecular basis of interferon-gamma and lipopolysaccharide enhancement of phagocyte respiratory burst capability. Studies on the gene expression of several NADPH oxidase components. J Biol Chem (1990) 265:20241–6.
122. Martin E, Nathan C, Xie QW. Role of interferon regulatory factor 1 in induction of nitric oxide synthase. J Exp Med (1994) 180:977–84. doi:10.1084/jem.180.3.977
123. Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol (1997) 15:563–91. doi:10.1146/annurev.immunol.15.1.563
124. Ihle JN, Kerr IM. Jaks and Stats in signaling by the cytokine receptor superfamily. Trends Genet (1995) 11:69–74. doi:10.1016/S0168-9525(00)89000-9
125. Darnell JE Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science (1994) 264:1415–21. doi:10.1126/science.8197455
126. Takaoka A, Mitani Y, Suemori H, Sato M, Yokochi T, Noguchi S, et al. Cross talk between interferon-gamma and -alpha/beta signaling components in caveolar membrane domains. Science (2000) 288:2357–60. doi:10.1126/science.288.5475.2357
127. Bluyssen HA, Muzaffar R, Vlieststra RJ, Van Der Made AC, Leung S, Stark GR, et al. Combinatorial association and abundance of components of interferon-stimulated gene factor 3 dictate the selectivity of interferon responses. Proc Natl Acad Sci U S A (1995) 92:5645–9. doi:10.1073/pnas.92.12.5645
128. Matsumoto M, Tanaka N, Harada H, Kimura T, Yokochi T, Kitagawa M, et al. Activation of the transcription factor ISGF3 by interferon-gamma. Biol Chem (1999) 380:699–703. doi:10.1515/BC.1999.087
129. Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell Microbiol (2006) 8:907–22. doi:10.1111/j.1462-5822.2006.00716.x
130. Young HA, Hardy KJ. Role of interferon-gamma in immune cell regulation. J Leukoc Biol (1995) 58:373–81. doi:10.1002/jlb.58.4.373
131. Zou J, Yoshiura Y, Dijkstra JM, Sakai M, Ototake M, Secombes C. Identification of an interferon gamma homologue in Fugu, Takifugu rubripes. Fish Shellfish Immunol (2004) 17:403–9. doi:10.1016/j.fsi.2004.04.015
132. Igawa D, Sakai M, Savan R. An unexpected discovery of two interferon gamma-like genes along with interleukin (IL)-22 and -26 from teleost: IL-22 and -26 genes have been described for the first time outside mammals. Mol Immunol (2006) 43:999–1009. doi:10.1016/j.molimm.2005.05.009
133. Milev-Milovanovic I, Long S, Wilson M, Bengten E, Miller NW, Chinchar VG. Identification and expression analysis of interferon gamma genes in channel catfish. Immunogenetics (2006) 58:70–80. doi:10.1007/s00251-006-0081-x
134. Zou J, Tafalla C, Truckle J, Secombes CJ. Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J Immunol (2007) 179:3859–71. doi:10.4049/jimmunol.179.6.3859
135. Zou J, Secombes CJ. Teleost fish interferons and their role in immunity. Dev Comp Immunol (2011) 35:1376–87. doi:10.1016/j.dci.2011.07.001
136. Zou J, Carrington A, Collet B, Dijkstra JM, Yoshiura Y, Bols N, et al. Identification and bioactivities of IFN-gamma in rainbow trout Oncorhynchus mykiss: the first Th1-type cytokine characterized functionally in fish. J Immunol (2005) 175:2484–94. doi:10.4049/jimmunol.175.4.2484
137. Wang T, Holland JW, Carrington A, Zou J, Secombes CJ. Molecular and functional characterization of IL-15 in rainbow trout Oncorhynchus mykiss: a potent inducer of IFN-gamma expression in spleen leukocytes. J Immunol (2007) 179:1475–88. doi:10.4049/jimmunol.179.3.1475
138. Grayfer L, Belosevic M. Molecular characterization, expression and functional analysis of goldfish (Carassius aurutus L.) interferon gamma. Dev Comp Immunol (2009) 33:235–46. doi:10.1016/j.dci.2008.09.001
139. Arts JA, Tijhaar EJ, Chadzinska M, Savelkoul HF, Verburg-Van Kemenade BM. Functional analysis of carp interferon-gamma: evolutionary conservation of classical phagocyte activation. Fish Shellfish Immunol (2010) 29:793–802. doi:10.1016/j.fsi.2010.07.010
140. Ruddle NH. Activation of human polymorphonuclear neutrophil functions by interferon-gamma and tumor necrosis factors. J Immunol (1986) 136:2335–6.
141. Gupta JW, Kubin M, Hartman L, Cassatella M, Trinchieri G. Induction of expression of genes encoding components of the respiratory burst oxidase during differentiation of human myeloid cell lines induced by tumor necrosis factor and gamma-interferon. Cancer Res (1992) 52:2530–7.
142. Yang S, Li Q, Mu Y, Ao J, Chen X. Functional activities of interferon gamma in large yellow croaker Larimichthys crocea. Fish Shellfish Immunol (2017) 70:545–52. doi:10.1016/j.fsi.2017.09.051
143. Ruan BY, Chen SN, Hou J, Huang B, Laghari ZA, Li L, et al. Two type II IFN members, IFN-gamma and IFN-gamma related (rel), regulate differentially IRF1 and IRF11 in zebrafish. Fish Shellfish Immunol (2017) 65:103–10. doi:10.1016/j.fsi.2017.03.054
144. Xiang Y, Liu W, Jia P, Li Y, Jin Y, Chen L, et al. Molecular characterization and expression analysis of interferon-gamma in black seabream Acanthopagrus schlegelii. Fish Shellfish Immunol (2017) 70:140–8. doi:10.1016/j.fsi.2017.08.046
145. Savan R, Ravichandran S, Collins JR, Sakai M, Young HA. Structural conservation of interferon gamma among vertebrates. Cytokine Growth Factor Rev (2009) 20:115–24. doi:10.1016/j.cytogfr.2009.02.006
146. Stolte EH, Savelkoul HF, Wiegertjes G, Flik G, Lidy Verburg-Van Kemenade BM. Differential expression of two interferon-gamma genes in common carp (Cyprinus carpio L.). Dev Comp Immunol (2008) 32:1467–81. doi:10.1016/j.dci.2008.06.012
147. Sieger D, Stein C, Neifer D, Van Der Sar AM, Leptin M. The role of gamma interferon in innate immunity in the zebrafish embryo. Dis Model Mech (2009) 2:571–81. doi:10.1242/dmm.003509
148. Grayfer L, Garcia EG, Belosevic M. Comparison of macrophage antimicrobial responses induced by type II interferons of the goldfish (Carassius auratus L.). J Biol Chem (2010) 285:23537–47. doi:10.1074/jbc.M109.096925
149. Yabu T, Toda H, Shibasaki Y, Araki K, Yamashita M, Anzai H, et al. Antiviral protection mechanisms mediated by ginbuna crucian carp interferon gamma isoforms 1 and 2 through two distinct interferon gamma-receptors. J Biochem (2011) 150:635–48. doi:10.1093/jb/mvr108
150. Shibasaki Y, Yabu T, Araki K, Mano N, Shiba H, Moritomo T, et al. Peculiar monomeric interferon gammas, IFNgammarel 1 and IFNgammarel 2, in ginbuna crucian carp. FEBS J (2014) 281:1046–56. doi:10.1111/febs.12666
151. Gao Q, Nie P, Thompson KD, Adams A, Wang T, Secombes CJ, et al. The search for the IFN-gamma receptor in fish: functional and expression analysis of putative binding and signalling chains in rainbow trout Oncorhynchus mykiss. Dev Comp Immunol (2009) 33:920–31. doi:10.1016/j.dci.2009.03.001
152. Farrar MA, Fernandez-Luna J, Schreiber RD. Identification of two regions within the cytoplasmic domain of the human interferon-gamma receptor required for function. J Biol Chem (1991) 266:19626–35.
153. Greenlund AC, Farrar MA, Viviano BL, Schreiber RD. Ligand-induced IFN gamma receptor tyrosine phosphorylation couples the receptor to its signal transduction system (p91). EMBO J (1994) 13:1591–600.
154. Greenlund AC, Morales MO, Viviano BL, Yan H, Krolewski J, Schreiber RD. Stat recruitment by tyrosine-phosphorylated cytokine receptors: an ordered reversible affinity-driven process. Immunity (1995) 2:677–87. doi:10.1016/1074-7613(95)90012-8
155. Grayfer L, Belosevic M. Molecular characterization of novel interferon gamma receptor 1 isoforms in zebrafish (Danio rerio) and goldfish (Carassius auratus L.). Mol Immunol (2009) 46:3050–9. doi:10.1016/j.molimm.2009.06.004
156. Aggad D, Stein C, Sieger D, Mazel M, Boudinot P, Herbomel P, et al. In vivo analysis of Ifn-γ1 and Ifn-γ2 signaling in zebrafish. J Immunol (2010) 185:6774–82. doi:10.4049/jimmunol.1000549
157. Ward AE, Rosenthal BM. Evolutionary responses of innate immunity to adaptive immunity. Infect Genet Evol (2014) 21:492–6. doi:10.1016/j.meegid.2013.12.021
158. Young JD, Liu CC, Butler G, Cohn ZA, Galli SJ. Identification, purification, and characterization of a mast cell-associated cytolytic factor related to tumor necrosis factor. Proc Natl Acad Sci U S A (1987) 84:9175–9. doi:10.1073/pnas.84.24.9175
159. Warner SJ, Libby P. Human vascular smooth muscle cells. Target for and source of tumor necrosis factor. J Immunol (1989) 142:100–9.
160. Dubravec DB, Spriggs DR, Mannick JA, Rodrick ML. Circulating human peripheral blood granulocytes synthesize and secrete tumor necrosis factor alpha. Proc Natl Acad Sci U S A (1990) 87:6758–61. doi:10.1073/pnas.87.17.6758
161. Carswell EA, Old LJ, Kassel RL, Green S, Fiore N, Williamson B. An endotoxin-induced serum factor that causes necrosis of tumors. Proc Natl Acad Sci U S A (1975) 72:3666–70. doi:10.1073/pnas.72.9.3666
162. Ming WJ, Bersani L, Mantovani A. Tumor necrosis factor is chemotactic for monocytes and polymorphonuclear leukocytes. J Immunol (1987) 138:1469–74.
163. Yonemaru M, Stephens KE, Ishizaka A, Zheng H, Hogue RS, Crowley JJ, et al. Effects of tumor necrosis factor on PMN chemotaxis, chemiluminescence, and elastase activity. J Lab Clin Med (1989) 114:674–81.
164. Klebanoff SJ, Vadas MA, Harlan JM, Sparks LH, Gamble JR, Agosti JM, et al. Stimulation of neutrophils by tumor necrosis factor. J Immunol (1986) 136:4220–5.
165. Lohmann-Matthes ML, Luttig B, Hockertz S. Involvement of membrane-associated TNF in the killing of Leishmania donovani parasites by macrophages. Behring Inst Mitt (1991) 88:125–32.
166. Van Strijp JA, Van Der Tol ME, Miltenburg LA, Van Kessel KP, Verhoef J. Tumour necrosis factor triggers granulocytes to internalize complement-coated virus particles. Immunology (1991) 73:77–82.
167. Ding AH, Nathan CF, Stuehr DJ. Release of reactive nitrogen intermediates and reactive oxygen intermediates from mouse peritoneal macrophages. Comparison of activating cytokines and evidence for independent production. J Immunol (1988) 141:2407–12.
168. Neumann M, Kownatzki E. The effect of adherence on the generation of reactive oxygen species by human neutrophilic granulocytes. Agents Actions (1989) 26:183–5. doi:10.1007/BF02126602
169. Schirren CG, Scharffetter K, Hein R, Braun-Falco O, Krieg T. Tumor necrosis factor alpha induces invasiveness of human skin fibroblasts in vitro. J Invest Dermatol (1990) 94:706–10. doi:10.1111/1523-1747.ep12876280
170. Camussi G, Bussolino F, Salvidio G, Baglioni C. Tumor necrosis factor/cachectin stimulates peritoneal macrophages, polymorphonuclear neutrophils, and vascular endothelial cells to synthesize and release platelet-activating factor. J Exp Med (1987) 166:1390–404. doi:10.1084/jem.166.5.1390
171. Hajjar KA, Hajjar DP, Silverstein RL, Nachman RL. Tumor necrosis factor-mediated release of platelet-derived growth factor from cultured endothelial cells. J Exp Med (1987) 166:235–45. doi:10.1084/jem.166.1.235
172. Li J, Zhang M, Rui YC. Tumor necrosis factor mediated release of platelet-derived growth factor from bovine cerebral microvascular endothelial cells. Zhongguo Yao Li Xue Bao (1997) 18:133–6.
173. Kriegler M, Perez C, Defay K, Albert I, Lu SD. A novel form of TNF/cachectin is a cell surface cytotoxic transmembrane protein: ramifications for the complex physiology of TNF. Cell (1988) 53:45–53. doi:10.1016/0092-8674(88)90486-2
174. Perez C, Albert I, Defay K, Zachariades N, Gooding L, Kriegler M. A nonsecretable cell surface mutant of tumor necrosis factor (TNF) kills by cell-to-cell contact. Cell (1990) 63:251–8. doi:10.1016/0092-8674(90)90158-B
175. Moss ML, Jin SL, Becherer JD, Bickett DM, Burkhart W, Chen WJ, et al. Structural features and biochemical properties of TNF-alpha converting enzyme (TACE). J Neuroimmunol (1997) 72:127–9. doi:10.1016/S0165-5728(96)00180-4
176. Eck MJ, Beutler B, Kuo G, Merryweather JP, Sprang SR. Crystallization of trimeric recombinant human tumor necrosis factor (cachectin). J Biol Chem (1988) 263:12816–9.
177. Banner DW, D’arcy A, Janes W, Gentz R, Schoenfeld HJ, Broger C, et al. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell (1993) 73:431–45. doi:10.1016/0092-8674(93)90132-A
178. Laing KJ, Wang T, Zou J, Holland J, Hong S, Bols N, et al. Cloning and expression analysis of rainbow trout Oncorhynchus mykiss tumour necrosis factor-alpha. Eur J Biochem (2001) 268:1315–22. doi:10.1046/j.1432-1327.2001.01996.x
179. Nguyen-Chi M, Laplace-Builhe B, Travnickova J, Luz-Crawford P, Tejedor G, Phan QT, et al. Identification of polarized macrophage subsets in zebrafish. Elife (2015) 4:e07288. doi:10.7554/eLife.07288
180. Ronza P, Losada AP, Villamarin A, Bermudez R, Quiroga MI. Immunolocalization of tumor necrosis factor alpha in turbot (Scophthalmus maximus, L.) tissues. Fish Shellfish Immunol (2015) 45:470–6. doi:10.1016/j.fsi.2015.04.032
181. Hirono I, Nam BH, Kurobe T, Aoki T. Molecular cloning, characterization, and expression of TNF cDNA and gene from Japanese flounder Paralychthys olivaceus. J Immunol (2000) 165:4423–7. doi:10.4049/jimmunol.165.8.4423
182. Nascimento DS, Pereira PJ, Reis MI, Do Vale A, Zou J, Silva MT, et al. Molecular cloning and expression analysis of sea bass (Dicentrarchus labrax L.) tumor necrosis factor-alpha (TNF-alpha). Fish Shellfish Immunol (2007) 23:701–10. doi:10.1016/j.fsi.2007.02.003
183. Ordas MC, Costa MM, Roca FJ, Lopez-Castejon G, Mulero V, Meseguer J, et al. Turbot TNFalpha gene: molecular characterization and biological activity of the recombinant protein. Mol Immunol (2007) 44:389–400. doi:10.1016/j.molimm.2006.02.028
184. Grayfer L, Walsh JG, Belosevic M. Characterization and functional analysis of goldfish (Carassius auratus L.) tumor necrosis factor-alpha. Dev Comp Immunol (2008) 32:532–43. doi:10.1016/j.dci.2007.09.009
185. Kadowaki T, Harada H, Sawada Y, Kohchi C, Soma G, Takahashi Y, et al. Two types of tumor necrosis factor-alpha in bluefin tuna (Thunnus orientalis) genes: molecular cloning and expression profile in response to several immunological stimulants. Fish Shellfish Immunol (2009) 27:585–94. doi:10.1016/j.fsi.2008.12.006
186. Lam FW, Wu SY, Lin SJ, Lin CC, Chen YM, Wang HC, et al. The expression of two novel orange-spotted grouper (Epinephelus coioides) TNF genes in peripheral blood leukocytes, various organs, and fish larvae. Fish Shellfish Immunol (2011) 30:618–29. doi:10.1016/j.fsi.2010.12.011
187. Zhang A, Chen D, Wei H, Du L, Zhao T, Wang X, et al. Functional characterization of TNF-alpha in grass carp head kidney leukocytes: induction and involvement in the regulation of NF-kappaB signaling. Fish Shellfish Immunol (2012) 33:1123–32. doi:10.1016/j.fsi.2012.08.029
188. Hong S, Li R, Xu Q, Secombes CJ, Wang T. Two types of TNF-α exist in teleost fish: phylogeny, expression, and bioactivity analysis of type-II TNF-α3 in rainbow trout Oncorhynchus mykiss. J Immunol (2013) 191:5959–72. doi:10.4049/jimmunol.1301584
189. Zou J, Peddie S, Scapigliati G, Zhang Y, Bols NC, Ellis AE, et al. Functional characterisation of the recombinant tumor necrosis factors in rainbow trout, Oncorhynchus mykiss. Dev Comp Immunol (2003) 27:813–22. doi:10.1016/S0145-305X(03)00077-6
190. Forlenza M, Fink IR, Raes G, Wiegertjes GF. Heterogeneity of macrophage activation in fish. Dev Comp Immunol (2011) 35:1246–55. doi:10.1016/j.dci.2011.03.008
191. Garcia-Castillo J, Chaves-Pozo E, Olivares P, Pelegrin P, Meseguer J, Mulero V. The tumor necrosis factor alpha of the bony fish seabream exhibits the in vivo proinflammatory and proliferative activities of its mammalian counterparts, yet it functions in a species-specific manner. Cell Mol Life Sci (2004) 61:1331–40. doi:10.1007/s00018-004-4068-1
192. Praveen K, Evans DL, Jaso-Friedmann L. Constitutive expression of tumor necrosis factor-alpha in cytotoxic cells of teleosts and its role in regulation of cell-mediated cytotoxicity. Mol Immunol (2006) 43:279–91. doi:10.1016/j.molimm.2005.01.012
193. Uenobe M, Kohchi C, Yoshioka N, Yuasa A, Inagawa H, Morii K, et al. Cloning and characterization of a TNF-like protein of Plecoglossus altivelis (ayu fish). Mol Immunol (2007) 44:1115–22. doi:10.1016/j.molimm.2006.07.281
194. Roca FJ, Mulero I, Lopez-Munoz A, Sepulcre MP, Renshaw SA, Meseguer J, et al. Evolution of the inflammatory response in vertebrates: fish TNF-alpha is a powerful activator of endothelial cells but hardly activates phagocytes. J Immunol (2008) 181:5071–81. doi:10.4049/jimmunol.181.7.5071
195. Li MF, Zhang J. CsTNF1, a teleost tumor necrosis factor that promotes antibacterial and antiviral immune defense in a manner that depends on the conserved receptor binding site. Dev Comp Immunol (2016) 55:65–75. doi:10.1016/j.dci.2015.10.010
196. Kutyrev I, Cleveland B, Leeds T, Wiens GD. Proinflammatory cytokine and cytokine receptor gene expression kinetics following challenge with Flavobacterium psychrophilum in resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol (2016) 58:542–53. doi:10.1016/j.fsi.2016.09.053
197. Kutyrev I, Cleveland B, Leeds T, Wiens GD. Dataset of proinflammatory cytokine and cytokine receptor gene expression in rainbow trout (Oncorhynchus mykiss) measured using a novel GeXP multiplex, RT-PCR assay. Data Brief (2017) 11:192–6. doi:10.1016/j.dib.2017.02.014
198. Bobe J, Goetz FW. Molecular cloning and expression of a TNF receptor and two TNF ligands in the fish ovary. Comp Biochem Physiol B Biochem Mol Biol (2001) 129:475–81.
199. Grayfer L, Belosevic M. Molecular characterization of tumor necrosis factor receptors 1 and 2 of the goldfish (Carassius aurutus L.). Mol Immunol (2009) 46:2190–9. doi:10.1016/j.molimm.2009.04.016
200. Tartaglia LA, Ayres TM, Wong GH, Goeddel DV. A novel domain within the 55 kd TNF receptor signals cell death. Cell (1993) 74:845–53. doi:10.1016/0092-8674(93)90464-2
201. Zhang S, Zhang R, Ma T, Qiu X, Wang X, Zhang A, et al. Identification and functional characterization of tumor necrosis factor receptor 1 (TNFR1) of grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol (2016) 58:24–32. doi:10.1016/j.fsi.2016.09.003
202. Pennica D, Kohr WJ, Fendly BM, Shire SJ, Raab HE, Borchardt PE, et al. Characterization of a recombinant extracellular domain of the type 1 tumor necrosis factor receptor: evidence for tumor necrosis factor-alpha induced receptor aggregation. Biochemistry (1992) 31:1134–41. doi:10.1021/bi00119a023
203. Naismith JH, Brandhuber BJ, Devine TQ, Sprang SR. Seeing double: crystal structures of the type I TNF receptor. J Mol Recognit (1996) 9:113–7. doi:10.1002/(SICI)1099-1352(199603)9:2<113::AID-JMR253>3.0.CO;2-H
204. Naismith JH, Devine TQ, Kohno T, Sprang SR. Structures of the extracellular domain of the type I tumor necrosis factor receptor. Structure (1996) 4:1251–62. doi:10.1016/S0969-2126(96)00134-7
205. Chapman BS, Kuntz ID. Modeled structure of the 75-kDa neurotrophin receptor. Protein Sci (1995) 4:1696–707. doi:10.1002/pro.5560040905
206. Gong Y, Cao P, Yu HJ, Jiang T. Crystal structure of the neurotrophin-3 and p75NTR symmetrical complex. Nature (2008) 454:789–93. doi:10.1038/nature07089
207. Huang X, Huang Y, Cai J, Wei S, Gao R, Qin Q. Identification and characterization of a tumor necrosis factor receptor like protein encoded by Singapore grouper iridovirus. Virus Res (2013) 178:340–8. doi:10.1016/j.virusres.2013.09.023
208. Pontejo SM, Sanchez C, Martin R, Mulero V, Alcami A, Alejo A. An orphan viral TNF receptor superfamily member identified in lymphocystis disease virus. Virol J (2013) 10:188. doi:10.1186/1743-422X-10-188
209. Yi Y, Qi H, Yuan J, Wang R, Weng S, He J, et al. Functional characterization of viral tumor necrosis factor receptors encoded by cyprinid herpesvirus 3 (CyHV3) genome. Fish Shellfish Immunol (2015) 45:757–70. doi:10.1016/j.fsi.2015.05.035
210. Yu Y, Huang Y, Wei S, Li P, Zhou L, Ni S, et al. A tumour necrosis factor receptor-like protein encoded by Singapore grouper iridovirus modulates cell proliferation, apoptosis and viral replication. J Gen Virol (2016) 97:756–66. doi:10.1099/jgv.0.000379
211. Gruys E, Toussaint MJ, Niewold TA, Koopmans SJ. Acute phase reaction and acute phase proteins. J Zhejiang Univ Sci B (2005) 6:1045–56. doi:10.1631/jzus.2005.B1045
212. Ingenbleek Y, Young V. Transthyretin (prealbumin) in health and disease: nutritional implications. Annu Rev Nutr (1994) 14:495–533. doi:10.1146/annurev.nu.14.070194.002431
213. Kimura T, Nakayama K, Penninger J, Kitagawa M, Harada H, Matsuyama T, et al. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science (1994) 264:1921–4. doi:10.1126/science.8009222
214. Heinrich PC, Castell JV, Andus T. Interleukin-6 and the acute phase response. Biochem J (1990) 265:621–36. doi:10.1042/bj2650621
215. Van Miert AS. Pro-inflammatory cytokines in a ruminant model: pathophysiological, pharmacological, and therapeutic aspects. Vet Q (1995) 17:41–50. doi:10.1080/01652176.1995.9694530
216. Heinrich PC, Horn F, Graeve L, Dittrich E, Kerr I, Muller-Newen G, et al. Interleukin-6 and related cytokines: effect on the acute phase reaction. Z Ernahrungswiss (1998) 37(Suppl 1):43–9.
217. Le J, Reis LF, Vilcek J. Tumor necrosis factor and interleukin 1 can act as essential growth factors in a murine plasmacytoma line. Lymphokine Res (1988) 7:99–106.
218. Sehgal PB, Helfgott DC, Santhanam U, Tatter SB, Clarick RH, Ghrayeb J, et al. Regulation of the acute phase and immune responses in viral disease. Enhanced expression of the beta 2-interferon/hepatocyte-stimulating factor/interleukin 6 gene in virus-infected human fibroblasts. J Exp Med (1988) 167:1951–6. doi:10.1084/jem.167.6.1951
219. Castell JV, Gomez-Lechon MJ, David M, Andus T, Geiger T, Trullenque R, et al. Interleukin-6 is the major regulator of acute phase protein synthesis in adult human hepatocytes. FEBS Lett (1989) 242:237–9. doi:10.1016/0014-5793(89)80476-4
220. Schindler R, Mancilla J, Endres S, Ghorbani R, Clark SC, Dinarello CA. Correlations and interactions in the production of interleukin-6 (IL-6), IL-1, and tumor necrosis factor (TNF) in human blood mononuclear cells: IL-6 suppresses IL-1 and TNF. Blood (1990) 75:40–7.
221. Knolle P, Lohr H, Treichel U, Dienes HP, Lohse A, Schlaack J, et al. Parenchymal and nonparenchymal liver cells and their interaction in the local immune response. Z Gastroenterol (1995) 33:613–20.
222. Winkelhake JL, Vodicnik MJ, Taylor JL. Induction in rainbow trout of an acute phase (C-reactive) protein by chemicals of environmental concern. Comp Biochem Physiol C (1983) 74:55–8. doi:10.1016/0742-8413(83)90148-2
223. Ghosh S, Bhattacharya S. Elevation of C-reactive protein in serum of Channa punctatus as an indicator of water pollution. Indian J Exp Biol (1992) 30:736–7.
224. Liu Y, Iwasaki T, Watarai S, Kodama H. Effect of turpentine oil on C-reactive protein (CRP) production in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol (2004) 17:203–10. doi:10.1016/j.fsi.2004.03.003
225. Kovacevic N, Hagen MO, Xie J, Belosevic M. The analysis of the acute phase response during the course of Trypanosoma carassii infection in the goldfish (Carasius auratus L.). Fish Shellfish Immunol (2015) 53:112–22. doi:10.1016/j.dci.2015.06.009
226. Jensen LE, Hiney MP, Shields DC, Uhlar CM, Lindsay AJ, Whitehead AS. Acute phase proteins in salmonids: evolutionary analyses and acute phase response. J Immunol (1997) 158:384–92.
227. Kovacevic N, Belosevic M. Molecular and functional characterization of goldfish Carassius auratus L.) serum amyloid A. Fish Shellfis Immunol (2015) 47:942–53. doi:10.1016/j.fsi.2015.10.041
228. Nakanishi Y, Kodama H, Murai T, Mikami T, Izawa H. Activation of rainbow trout complement by C-reactive protein. Am J Vet Res (1991) 52:397–401.
229. Jorgensen JB, Lunde H, Jensen L, Whitehead AS, Robertsen B. Serum amyloid A transcription in Atlantic salmon (Salmo salar L.) hepatocytes is enhanced by stimulation with macrophage factors, recombinant human IL-1 beta, IL-6 and TNF alpha or bacterial lipopolysaccharide. Dev Comp Immunol (2000) 24:553–63. doi:10.1016/S0145-305X(00)00022-7
230. Lund V, Olafsen JA. Changes in serum concentration of a serum amyloid P-like pentraxin in Atlantic salmon, Salmo salar L., during infection and inflammation. Dev Comp Immunol (1999) 23:61–70. doi:10.1016/S0145-305X(98)00038-X
231. Gerwick L, Steinhauer R, Lapatra S, Sandell T, Ortuno J, Hajiseyedjavadi N, et al. The acute phase response of rainbow trout (Oncorhynchus mykiss) plasma proteins to viral, bacterial and fungal inflammatory agents. Fish Shellfish Immunol (2002) 12:229–42. doi:10.1006/fsim.2001.0367
232. Peatman E, Baoprasertkul P, Terhune J, Xu P, Nandi S, Kucuktas H, et al. Expression analysis of the acute phase response in channel catfish (Ictalurus punctatus) after infection with a Gram-negative bacterium. Dev Comp Immunol (2007) 31:1183–96. doi:10.1016/j.dci.2007.03.003
233. Lin B, Chen S, Cao Z, Lin Y, Mo D, Zhang H, et al. Acute phase response in zebrafish upon Aeromonas salmonicida and Staphylococcus aureus infection: striking similarities and obvious differences with mammals. Mol Immunol (2007) 44:295–301. doi:10.1016/j.molimm.2006.03.001
234. Mastellos D, Morikis D, Isaacs SN, Holland MC, Strey CW, Lambris JD. Complement: structure, functions, evolution, and viral molecular mimicry. Immunol Res (2003) 27:367–86. doi:10.1385/IR:27:2-3:367
235. Markiewski MM, Lambris JD. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am J Pathol (2007) 171:715–27. doi:10.2353/ajpath.2007.070166
236. Nonaka M. Origin and evolution of the complement system. Curr Top Microbiol Immunol (2000) 248:37–50.
237. Sunyer JO, Zarkadis IK, Sahu A, Lambris JD. Multiple forms of complement C3 in trout that differ in binding to complement activators. Proc Natl Acad Sci U S A (1996) 93:8546–51. doi:10.1073/pnas.93.16.8546
238. Sunyer JO, Tort L, Lambris JD. Diversity of the third form of complement, C3, in fish: functional characterization of five forms of C3 in the diploid fish Sparus aurata. Biochem J (1997) 326(Pt 3):877–81. doi:10.1042/bj3260877
239. Sunyer JO, Tort L, Lambris JD. Structural C3 diversity in fish: characterization of five forms of C3 in the diploid fish Sparus aurata. J Immunol (1997) 158:2813–21.
240. Kato Y, Nakao M, Shimizu M, Wariishi H, Yano T. Purification and functional assessment of C3a, C4a and C5a of the common carp (Cyprinus carpio) complement. Dev Comp Immunol (2004) 28:901–10. doi:10.1016/j.dci.2004.01.006
241. Rotllant J, Parra D, Peters R, Boshra H, Sunyer JO. Generation, purification and functional characterization of three C3a anaphylatoxins in rainbow trout: role in leukocyte chemotaxis and respiratory burst. Dev Comp Immunol (2004) 28:815–28. doi:10.1016/j.dci.2003.11.001
242. Li J, Peters R, Lapatra S, Vazzana M, Sunyer JO. Anaphylatoxin-like molecules generated during complement activation induce a dramatic enhancement of particle uptake in rainbow trout phagocytes. Dev Comp Immunol (2004) 28:1005–21. doi:10.1016/j.dci.2004.03.004
243. Sunyer JO, Boshra H, Li J. Evolution of anaphylatoxins, their diversity and novel roles in innate immunity: insights from the study of fish complement. Vet Immunol Immunopathol (2005) 108:77–89. doi:10.1016/j.vetimm.2005.07.009
244. Alberdi F Jr, Alderton MR, Coloe PJ, Smith SC. Characterization of immunorelated peptides to porcidin P1. Immunol Cell Biol (1995) 73:505–10. doi:10.1038/icb.1995.80
245. Alberdi F Jr, Alderton MR, Korolik V, Coloe PJ, Smith SC. Antibacterial proteins from porcine polymorphonuclear neutrophils. Immunol Cell Biol (1995) 73:38–43. doi:10.1038/icb.1995.6
246. Kaplan DH, Greenlund AC, Tanner JW, Shaw AS, Schreiber RD. Identification of an interferon-gamma receptor alpha chain sequence required for JAK-1 binding. J Biol Chem (1996) 271:9–12. doi:10.1074/jbc.271.1.9
247. Ganz T, Lehrer RI. Antimicrobial peptides of leukocytes. Curr Opin Hematol (1997) 4:53–8. doi:10.1097/00062752-199704010-00009
248. Borelli V, Banfi E, Perrotta MG, Zabucchi G. Myeloperoxidase exerts microbicidal activity against Mycobacterium tuberculosis. Infect Immun (1999) 67:4149–52.
249. Sorensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, Hiemstra PS, et al. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood (2001) 97:3951–9. doi:10.1182/blood.V97.12.3951
250. Briggs RT, Drath DB, Karnovsky ML, Karnovsky MJ. Localization of NADH oxidase on the surface of human polymorphonuclear leukocytes by a new cytochemical method. J Cell Biol (1975) 67:566–86. doi:10.1083/jcb.67.3.566
251. DeLeo FR, Quinn MT. Assembly of the phagocyte NADPH oxidase: molecular interaction of oxidase proteins. J Leukoc Biol (1996) 60:677–91. doi:10.1002/jlb.60.6.677
252. El-Benna J, Dang PM-C, Gougerot-Pocidalo M-A. Priming of the neutrophil NADPH oxidase activation: role of p47phox phosphorylation and NOX2 mobilization to the plasma membrane. Semin Immunopathol (2008) 30:279–89. doi:10.1007/s00281-008-0118-3
253. Parkos CA, Allen RA, Cochrane CG, Jesaitis AJ. Purified cytochrome b from human granulocyte plasma membrane is comprised of two polypeptides with relative molecular weights of 91,000 and 22,000. J Clin Invest (1987) 80:732–42. doi:10.1172/JCI113128
254. Parkos CA, Allen RA, Cochrane CG, Jesaitis AJ. The quaternary structure of the plasma membrane b-type cytochrome of human granulocytes. Biochim Biophys Acta (1988) 932:71–83. doi:10.1016/0005-2728(88)90140-5
255. Parkos CA, Dinauer MC, Walker LE, Allen RA, Jesaitis AJ, Orkin SH. Primary structure and unique expression of the 22-kilodalton light chain of human neutrophil cytochrome b. Proc Natl Acad Sci U S A (1988) 85:3319–23. doi:10.1073/pnas.85.10.3319
256. Volpp BD, Nauseef WM, Donelson JE, Moser DR, Clark RA. Cloning of the cDNA and functional expression of the 47-kilodalton cytosolic component of human neutrophil respiratory burst oxidase. Proc Natl Acad Sci U S A (1989) 86:7195–9. doi:10.1073/pnas.86.18.7195
257. Leto TL, Lomax KJ, Volpp BD, Nunoi H, Sechler JM, Nauseef WM, et al. Cloning of a 67-kD neutrophil oxidase factor with similarity to a noncatalytic region of p60c-src. Science (1990) 248:727–30. doi:10.1126/science.1692159
258. Tsunawaki S, Mizunari H, Nagata M, Tatsuzawa O, Kuratsuji T. A novel cytosolic component, p40phox, of respiratory burst oxidase associates with p67phox and is absent in patients with chronic granulomatous disease who lack p67phox. Biochem Biophys Res Commun (1994) 199:1378–87. doi:10.1006/bbrc.1994.1383
259. Jorgensen JB, Robertsen B. Yeast beta-glucan stimulates respiratory burst activity of Atlantic salmon (Salmo salar L.) macrophages. Dev Comp Immunol (1995) 19:43–57. doi:10.1016/0145-305X(94)00045-H
260. Stafford JL, Galvez F, Goss GG, Belosevic M. Induction of nitric oxide and respiratory burst response in activated goldfish macrophages requires potassium channel activity. Dev Comp Immunol (2002) 26:445–59. doi:10.1016/S0145-305X(01)00087-8
261. Sepulcre MP, Lopez-Castejon G, Meseguer J, Mulero V. The activation of gilthead seabream professional phagocytes by different PAMPs underlines the behavioural diversity of the main innate immune cells of bony fish. Mol Immunol (2007) 44:2009–16. doi:10.1016/j.molimm.2006.09.022
262. Boltana S, Donate C, Goetz FW, Mackenzie S, Balasch JC. Characterization and expression of NADPH oxidase in LPS-, poly(I:C)- and zymosan-stimulated trout (Oncorhynchus mykiss W.) macrophages. Fish Shellfish Immunol (2009) 26:651–61. doi:10.1016/j.fsi.2008.11.011
263. Sharp GJE, Secombes CJ. The role of reactive oxygen species in the killing of the bacterial fish pathogen Aeromonas salmonicida by rainbow trout macrophages. Fish Shellfish Immunol (1993) 3:119–29. doi:10.1006/fsim.1993.1013
264. Ardó L, Jeney Z, Adams A, Jeney G. Immune responses of resistant and sensitive common carp families following experimental challenge with Aeromonas hydrophila. Fish Shellfish Immunol (2010) 29:111–6. doi:10.1016/j.fsi.2010.02.029
265. Hodgkinson JW, Ge JQ, Grayfer L, Stafford J, Belosevic M. Analysis of the immune response in infections of the goldfish (Carassius auratus L.) with Mycobacterium marinum. Dev Comp Immunol (2012) 38:456–65. doi:10.1016/j.dci.2012.07.006
266. Kim MS, Hwang YJ, Yoon KJ, Zenke K, Nam YK, Kim SK, et al. Molecular cloning of rock bream (Oplegnathus fasciatus) tumor necrosis factor-α and its effect on the respiratory burst activity of phagocytes. Fish Shellfish Immunol (2009) 27:618–24. doi:10.1016/j.fsi.2009.07.007
267. Taylor MW, Feng GS. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J (1991) 5:2516–22. doi:10.1096/fasebj.5.11.1907934
268. Wang X-F, Wang H-S, Wang H, Zhang F, Wang K-F, Guo Q, et al. The role of indoleamine 2,3-dioxygenase (IDO) in immune tolerance: Focus on macrophage polarization of THP-1 cells. Cell Immunol (2014) 289:42–8. doi:10.1016/j.cellimm.2014.02.005
269. Grohmann U, Bronte V. Control of immune response by amino acid metabolism. Immunol Rev (2010) 236:243–64. doi:10.1111/j.1600-065X.2010.00915.x
270. Yoshida R, Imanishi J, Oku T, Kishida T, Hayaishi O. Induction of pulmonary indoleamine 2,3-dioxygenase by interferon. Proc Natl Acad Sci U S A (1981) 78:129–32. doi:10.1073/pnas.78.1.129
271. Werner ER, Werner-Felmayer G, Fuchs D, Hausen A, Reibnegger G, Wachter H. Parallel induction of tetrahydrobiopterin biosynthesis and indoleamine 2,3-dioxygenase activity in human cells and cell lines by interferon-gamma. Biochem J (1989) 262:861–6. doi:10.1042/bj2620861
272. Werner-Felmayer G, Werner ER, Fuchs D, Hausen A, Reibnegger G, Wachter H. Induction of indoleamine 2,3-dioxygenase in human cells in vitro. In: Schwarcz R, Young SN, Brown RR, editors. Kynurenine and Serotonin Pathways. New York: Springer (1991). p. 505–9.
273. Hayaishi O. Utilization of superoxide anion by indoleamine oxygenase-catalyzed tryptophan and indoleamine oxidation. In: Filippini GA, Costa CVL, Bertazzo A, editors. Recent Advances in Tryptophan Research. US: Springer (1996). p. 285–9.
274. Yuasa HJ, Takubo M, Takahashi A, Hasegawa T, Noma H, Suzuki T. Evolution of vertebrate indoleamine 2,3-dioxygenases. J Mol Evol (2007) 65:705–14. doi:10.1007/s00239-007-9049-1
275. Grayfer L, Hodgkinson JW, Belosevic M. Analysis of the antimicrobial responses of primary phagocytes of the goldfish (Carassius auratus L.) against Mycobacterium marinum. Dev Comp Immunol (2011) 35:1146–58. doi:10.1016/j.dci.2011.04.007
276. John M, Qiao-Wen X, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol (1997) 15:323–50. doi:10.1146/annurev.immunol.15.1.323
277. Henard CA, Vázquez-Torres A. Nitric oxide and salmonella pathogenesis. Front Microbiol (2011) 2:84. doi:10.3389/fmicb.2011.00084
278. Saeij JP, Stet RJ, Groeneveld A, Verburg-Van Kemenade LB, Van Muiswinkel WB, Wiegertjes GF. Molecular and functional characterization of a fish inducible-type nitric oxide synthase. Immunogenetics (2000) 51:339–46. doi:10.1007/s002510050628
279. Joerink M, Ribeiro CMS, Stet RJM, Hermsen T, Savelkoul HFJ, Wiegertjes GF. Head kidney-derived macrophages of common carp (Cyprinus carpio L.) show plasticity and functional polarization upon differential stimulation. J Immunol (2006) 177:61–9. doi:10.4049/jimmunol.177.1.61
280. Stafford JL, Neumann NF, Belosevic M. Products of proteolytic cleavage of transferrin induce nitric oxide response of goldfish macrophages. Dev Comp Immunol (2001) 25:101–15. doi:10.1016/S0145-305X(00)00048-3
281. Stafford JL, Wilson EC, Belosevic M. Recombinant transferrin induces nitric oxide response in goldfish and murine macrophages. Fish Shellfish Immunol (2004) 17:171–85. doi:10.1016/j.fsi.2004.03.001
282. Tafalla C, Figueras A, Novoa B. Role of nitric oxide on the replication of viral haemorrhagic septicemia virus (VHSV), a fish rhabdovirus. Vet Immunol Immunopathol (1999) 72:249–56. doi:10.1016/S0165-2427(99)00109-9
283. Campos-Pérez JJ, Ellis AE, Secombes CJ. Toxicity of nitric oxide and peroxynitrite to bacterial pathogens of fish. Dis Aquat Organ (2000) 43:109–15. doi:10.3354/dao043109
284. Chettri JK, Raida MK, Kania PW, Buchmann K. Differential immune response of rainbow trout (Oncorhynchus mykiss) at early developmental stages (larvae and fry) against the bacterial pathogen Yersinia ruckeri. Dev Comp Immunol (2012) 36:463–74. doi:10.1016/j.dci.2011.08.014
285. Elks PM, Brizee S, Van Der Vaart M, Walmsley SR, Van Eeden FJ, Renshaw SA, et al. Hypoxia inducible factor signaling modulates susceptibility to mycobacterial infection via a nitric oxide dependent mechanism. PLoS Pathog (2013) 9:e1003789. doi:10.1371/journal.ppat.1003789
286. Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, Urdahl KB, et al. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature (2014) 505:218–22. doi:10.1038/nature12799
287. Elks PM, Van Der Vaart M, Van Hensbergen V, Schutz E, Redd MJ, Murayama E, et al. Mycobacteria counteract a TLR-mediated nitrosative defense mechanism in a zebrafish infection model. PLoS One (2014) 9:e100928. doi:10.1371/journal.pone.0100928
288. Iyengar R, Stuehr DJ, Marletta MA. Macrophage synthesis of nitrite, nitrate, and N-nitrosamines: precursors and role of the respiratory burst. Proc Natl Acad Sci U S A (1987) 84:6369–73. doi:10.1073/pnas.84.18.6369
289. Neumann NF, Belosevic M. Deactivation of primed respiratory burst response of goldfish macrophages by leukocyte-derived macrophage activating factor(s). Dev Comp Immunol (1996) 20:427–39. doi:10.1016/S0145-305X(96)00029-8
290. Neumann NF, Barreda DR, Belosevic M. Generation and functional analysis of distinct macrophage sub-populations from goldfish (Carassius auratus L.) kidney leukocyte cultures. Fish Shellfish Immunol (2000) 10:1–20. doi:10.1006/fsim.1999.0221
291. Martin JH, Edwards SW. Changes in mechanisms of monocyte/macrophage-mediated cytotoxicity during culture. Reactive oxygen intermediates are involved in monocyte-mediated cytotoxicity, whereas reactive nitrogen intermediates are employed by macrophages in tumor cell killing. J Immunol (1993) 150:3478–86.
292. Werner ER, Werner-Felmayer G. Substrate and cofactor requirements of indoleamine 2,3-dioxygenase in interferon-gamma-treated cells: utilization of oxygen rather than superoxide. Curr Drug Metab (2007) 8:201–3. doi:10.2174/138920007780362482
293. Yeung AW, Terentis AC, King NJ, Thomas SR. Role of indoleamine 2,3-dioxygenase in health and disease. Clin Sci (Lond) (2015) 129:601–72. doi:10.1042/CS20140392
294. Hirata F, Ohnishi T, Hayaishi O. Indoleamine 2,3-dioxygenase. Characterization and properties of enzyme. O2- complex. J Biol Chem (1977) 252:4637–42.
295. Taniguchi T, Sono M, Hirata F, Hayaishi O, Tamura M, Hayashi K, et al. Indoleamine 2,3-dioxygenase. Kinetic studies on the binding of superoxide anion and molecular oxygen to enzyme. J Biol Chem (1979) 254:3288–94.
296. Collins MA, Neafsey EJ, Cheng BY, Hurley-Gius K, Ung-Chhun NA, Pronger DA, et al. Endogenous analogs of N-methyl-4-phenyl-1,2,3,6-tetrahydropyridine: indoleamine derived tetrahydro-beta-carbolines as potential causative factors in Parkinson’s disease. Adv Neurol (1987) 45:179–82.
297. Christen S, Peterhans E, Stocker R. Antioxidant activities of some tryptophan metabolites: possible implication for inflammatory diseases. Proc Natl Acad Sci U S A (1990) 87:2506–10. doi:10.1073/pnas.87.7.2506
298. Melillo G, Cox GW, Radzioch D, Varesio L. Picolinic acid, a catabolite of L-tryptophan, is a costimulus for the induction of reactive nitrogen intermediate production in murine macrophages. J Immunol (1993) 150:4031–40.
299. Melillo G, Cox GW, Biragyn A, Sheffler LA, Varesio L. Regulation of nitric-oxide synthase mRNA expression by interferon-gamma and picolinic acid. J Biol Chem (1994) 269:8128–33.
300. Melillo G, Bosco MC, Musso T, Varesio L. Immunobiology of picolinic acid. Adv Exp Med Biol (1996) 398:135–41. doi:10.1007/978-1-4613-0381-7_22
301. Bosco MC, Rapisarda A, Massazza S, Melillo G, Young H, Varesio L. The tryptophan catabolite picolinic acid selectively induces the chemokines macrophage inflammatory protein-1 alpha and -1 beta in macrophages. J Immunol (2000) 164:3283–91. doi:10.4049/jimmunol.164.6.3283
302. Pastorino S, Carta L, Puppo M, Melillo G, Bosco MC, Varesio L. Picolinic acid- or desferrioxamine-inducible autocrine activation of macrophages engineered to produce IFNgamma: an approach for gene therapy. Gene Ther (2004) 11:560–8. doi:10.1038/sj.gt.3302217
303. Gopalakrishna R, Chen ZH, Gundimeda U. Nitric oxide and nitric oxide-generating agents induce a reversible inactivation of protein kinase C activity and phorbol ester binding. J Biol Chem (1993) 268:27180–5.
304. Thomas SR, Mohr D, Stocker R. Nitric oxide inhibits indoleamine 2,3-dioxygenase activity in interferon-gamma primed mononuclear phagocytes. J Biol Chem (1994) 269:14457–64.
305. Griscavage JM, Rogers NE, Sherman MP, Ignarro LJ. Inducible nitric oxide synthase from a rat alveolar macrophage cell line is inhibited by nitric oxide. J Immunol (1993) 151:6329–37.
306. Griscavage JM, Wilk S, Ignarro LJ. Serine and cysteine proteinase inhibitors prevent nitric oxide production by activated macrophages by interfering with transcription of the inducible NO synthase gene. Biochem Biophys Res Commun (1995) 215:721–9. doi:10.1006/bbrc.1995.2523
307. Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep (2014) 6:13. doi:10.12703/P6-13
308. Roberts SB, Langenau DM, Goetz FW. Cloning and characterization of prostaglandin endoperoxide synthase-1 and -2 from the brook trout ovary. Mol Cell Endocrinol (2000) 160:89–97. doi:10.1016/S0303-7207(99)00252-X
309. Gardiner MR, Daggett DF, Zon LI, Perkins AC. Zebrafish KLF4 is essential for anterior mesendoderm/pre-polster differentiation and hatching. Dev Dyn (2005) 234:992–6. doi:10.1002/dvdy.20571
310. Gardiner MR, Gongora MM, Grimmond SM, Perkins AC. A global role for zebrafish klf4 in embryonic erythropoiesis. Mech Dev (2007) 124:762–74. doi:10.1016/j.mod.2007.06.005
311. Martinez FO, Helming L, Milde R, Varin A, Melgert BN, Draijer C, et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood (2013) 121:e57–69. doi:10.1182/blood-2012-06-436212
312. Skjesol A, Liebe T, Iliev DB, Thomassen EI, Tollersrud LG, Sobhkhez M, et al. Functional conservation of suppressors of cytokine signaling proteins between teleosts and mammals: Atlantic salmon SOCS1 binds to JAK/STAT family members and suppresses type I and II IFN signaling. Dev Comp Immunol (2014) 45:177–89. doi:10.1016/j.dci.2014.02.009
313. Varela M, Diaz-Rosales P, Pereiro P, Forn-Cuni G, Costa MM, Dios S, et al. Interferon-induced genes of the expanded IFIT family show conserved antiviral activities in non-mammalian species. PLoS One (2014) 9:e100015. doi:10.1371/journal.pone.0100015
314. Ohtani M, Hayashi N, Hashimoto K, Nakanishi T, Dijkstra JM. Comprehensive clarification of two paralogous interleukin 4/13 loci in teleost fish. Immunogenetics (2008) 60:383–97. doi:10.1007/s00251-008-0299-x
315. Wang T, Secombes CJ. The evolution of IL-4 and IL-13 and their receptor subunits. Cytokine (2015) 75:8–13. doi:10.1016/j.cyto.2015.04.012
316. Wang T, Huang W, Costa MM, Martin SA, Secombes CJ. Two copies of the genes encoding the subunits of putative interleukin (IL)-4/IL-13 receptors, IL-4Rα, IL-13Rα1 and IL-13Rα2, have been identified in rainbow trout (Oncorhynchus mykiss) and have complex patterns of expression and modulation. Immunogenetics (2011) 63:235–53. doi:10.1007/s00251-010-0508-2
317. Zhu L-Y, Pan P-P, Fang W, Shao J-Z, Xiang L-X. Essential role of IL-4 and IL-4Rα interaction in adaptive immunity of zebrafish: insight into the origin of Th2-like regulatory mechanism in ancient vertebrates. J Immunol (2012) 188:5571–84. doi:10.4049/jimmunol.1102259
318. Lin A-F, Xiang L-X, Wang Q-L, Dong W-R, Gong Y-F, Shao J-Z. The DC-SIGN of zebrafish: insights into the existence of a CD209 homologue in a lower vertebrate and its involvement in adaptive immunity. J Immunol (2009) 183:7398–410. doi:10.4049/jimmunol.0803955
319. Hu Y-L, Xiang L-X, Shao J-Z. Identification and characterization of a novel immunoglobulin Z isotype in zebrafish: implications for a distinct B cell receptor in lower vertebrates. Mol Immunol (2010) 47:738–46. doi:10.1016/j.molimm.2009.10.010
320. Takizawa F, Koppang EO, Ohtani M, Nakanishi T, Hashimoto K, Fischer U, et al. Constitutive high expression of interleukin-4/13A and GATA-3 in gill and skin of salmonid fishes suggests that these tissues form Th2-skewed immune environments. Mol Immunol (2011) 48:1360–8. doi:10.1016/j.molimm.2011.02.014
321. Wang T, Johansson P, Abos B, Holt A, Tafalla C, Jiang Y, et al. First in-depth analysis of the novel Th2-type cytokines in salmonid fish reveals distinct patterns of expression and modulation but overlapping bioactivities. Oncotarget (2016) 7:10917–46. doi:10.18632/oncotarget.7295
322. Yang ZJ, Li CH, Chen J, Zhang H, Li MY, Chen J. Molecular characterization of an interleukin-4/13B homolog in grass carp (Ctenopharyngodon idella) and its role in fish against Aeromonas hydrophila infection. Fish Shellfish Immunol (2016) 57:136–47. doi:10.1016/j.fsi.2016.08.022
323. Hodgkinson JW, Fibke C, Belosevic M. Recombinant IL-4/13A and IL-4/13B induce arginase activity and down-regulate nitric oxide response of primary goldfish (Carassius auratus L.) macrophages. Dev Comp Immunol (2017) 67:377–84. doi:10.1016/j.dci.2016.08.014
324. Stocchi V, Wang T, Randelli E, Mazzini M, Gerdol M, Pallavicini A, et al. Evolution of Th2 responses: characterization of IL-4/13 in sea bass (Dicentrarchus labrax L.) and studies of expression and biological activity. Sci Rep (2017) 7:2240. doi:10.1038/s41598-017-02472-y
325. Yamaguchi T, Takizawa F, Fischer U, Dijkstra JM. Along the axis between Type 1 and Type 2 immunity; principles conserved in evolution from fish to mammals. Biology (Basel) (2015) 4:814–59. doi:10.3390/biology4040814
326. Green SJ, Crawford RM, Hockmeyer JT, Meltzer MS, Nacy CA. Leishmania major amastigotes initiate the L-arginine-dependent killing mechanism in IFN-gamma-stimulated macrophages by induction of tumor necrosis factor-alpha. J Immunol (1990) 145:4290–7.
327. Barksdale AR, Bernard AC, Maley ME, Gellin GL, Kearney PA, Boulanger BR, et al. Regulation of arginase expression by T-helper II cytokines and isoproterenol. Surgery (2004) 135:527–35. doi:10.1016/j.surg.2003.10.007
328. Mills CD. Macrophage arginine metabolism to ornithine/urea or nitric oxide/citrulline: a life or death issue. Crit Rev Immunol (2001) 21:399–425. doi:10.1615/CritRevImmunol.v21.i5.10
329. Morris SM. Recent advances in arginine metabolism: roles and regulation of the arginases. Br J Pharmacol (2009) 157:922–30. doi:10.1111/j.1476-5381.2009.00278.x
330. Munder M, Eichmann K, Morán JM, Centeno F, Soler G, Modolell M. Th1/Th2-regulated expression of arginase isoforms in murine macrophages and dendritic cells. J Immunol (1999) 163:3771–7.
331. Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol (2002) 169:2253–63. doi:10.4049/jimmunol.169.5.2253
332. Wright PA, Campbell A, Morgan RL, Rosenberger AG, Murray BW. Dogmas and controversies in the handling of nitrogenous wastes: expression of arginase Type I and II genes in rainbow trout: influence of fasting on liver enzyme activity and mRNA levels in juveniles. J Exp Biol (2004) 207:2033–42. doi:10.1242/jeb.00958
333. Wiegertjes GF, Wentzel AS, Spaink HP, Elks PM, Fink IR. Polarization of immune responses in fish: the ‘macrophages first’ point of view. Mol Immunol (2016) 69:146–56. doi:10.1016/j.molimm.2015.09.026
334. Ehrchen J, Steinmüller L, Barczyk K, Tenbrock K, Nacken W, Eisenacher M, et al. Glucocorticoids induce differentiation of a specifically activated, anti-inflammatory subtype of human monocytes. Blood (2007) 109:1265–74. doi:10.1182/blood-2006-02-001115
335. van de Garde MD, Martinez FO, Melgert BN, Hylkema MN, Jonkers RE, Hamann J. Chronic exposure to glucocorticoids shapes gene expression and modulates innate and adaptive activation pathways in macrophages with distinct changes in leukocyte attraction. J Immunol (2014) 192:1196–208. doi:10.4049/jimmunol.1302138
336. Torrecillas S, Montero D, Caballero MJ, Robaina L, Zamorano MJ, Sweetman J, et al. Effects of dietary concentrated mannan oligosaccharides supplementation on growth, gut mucosal immune system and liver lipid metabolism of European sea bass (Dicentrarchus labrax) juveniles. Fish Shellfish Immunol (2015) 42:508–16. doi:10.1016/j.fsi.2014.11.033
337. Wang R, Belosevic M. The in vitro effects of estradiol and cortisol on the function of a long-term goldfish macrophage cell line. Dev Comp Immunol (1995) 19:327–36. doi:10.1016/0145-305X(95)00018-O
338. Ferrante CJ, Leibovich SJ. Regulation of macrophage polarization and wound healing. Adv Wound Care (New Rochelle) (2012) 1:10–6. doi:10.1089/wound.2011.0307
339. Grayfer L, Belosevic M. Identification and molecular characterization of the interleukin-10 receptor 1 of the zebrafish (Danio rerio) and the goldfish (Carassius auratus L.). Dev Comp Immunol (2012) 36:408–17. doi:10.1016/j.dci.2011.08.006
340. He Wei SW. Molecular characterization, 3D modeling of grass carp interleukin-10 receptor 1 (IL10R1). Engineering (2013) 5:214–9. doi:10.4236/eng.2013.510B045
341. Monte MM, Wang T, Collet B, Zou J, Secombes CJ. Molecular characterisation of four class 2 cytokine receptor family members in rainbow trout, Oncorhynchus mykiss. Dev Comp Immunol (2015) 48:43–54. doi:10.1016/j.dci.2014.08.012
342. Lehner PJ, Cresswell P. Recent developments in MHC-class-I-mediated antigen presentation. Curr Opin Immunol (2004) 16:82–9. doi:10.1016/j.coi.2003.11.012
343. Brode S, Macary PA. Cross-presentation: dendritic cells and macrophages bite off more than they can chew! Immunology (2004) 112:345–51. doi:10.1111/j.1365-2567.2004.01920.x
344. Roche PA, Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol (2015) 15:203–16. doi:10.1038/nri3818
345. Chancellor A, Gadola SD, Mansour S. The versatility of the CD1 lipid antigen presentation pathway. Immunology (2018). doi:10.1111/imm.12912
346. Lennartz M, Drake J. Molecular mechanisms of macrophage toll-like receptor-Fc receptor synergy. F1000Res (2018) 7:21. doi:10.12688/f1000research.12679.1
347. Sever L, Vo NT, Bols NC, Dixon B. Expression of tapasin in rainbow trout tissues and cell lines and up regulation in a monocyte/macrophage cell line (RTS11) by a viral mimic and viral infection. Dev Comp Immunol (2014) 44:86–93. doi:10.1016/j.dci.2013.11.019
348. Sever L, Vo NT, Lumsden J, Bols NC, Dixon B. Induction of rainbow trout MH class I and accessory proteins by viral haemorrhagic septicaemia virus. Mol Immunol (2014) 59:154–62. doi:10.1016/j.molimm.2014.02.001
349. Sever L, Vo NTK, Bols NC, Dixon B. Tapasin’s protein interactions in the rainbow trout peptide-loading complex. Dev Comp Immunol (2018) 81:262–70. doi:10.1016/j.dci.2017.12.015
350. Tsukamoto K, Miura F, Fujito NT, Yoshizaki G, Nonaka M. Long-lived dichotomous lineages of the proteasome subunit beta type 8 (PSMB8) gene surviving more than 500 million years as alleles or paralogs. Mol Biol Evol (2012) 29:3071–9. doi:10.1093/molbev/mss113
351. McConnell SC, Hernandez KM, Wcisel DJ, Kettleborough RN, Stemple DL, Yoder JA, et al. Alternative haplotypes of antigen processing genes in zebrafish diverged early in vertebrate evolution. Proc Natl Acad Sci U S A (2016) 113:E5014–23. doi:10.1073/pnas.1607602113
352. Grimholt U. Whole genome duplications have provided teleosts with many roads to peptide loaded MHC class I molecules. BMC Evol Biol (2018) 18:25. doi:10.1186/s12862-018-1138-9
353. Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrom M, Gregers TF, et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature (2011) 477:207–10. doi:10.1038/nature10342
354. Star B, Jentoft S. Why does the immune system of Atlantic cod lack MHC II? Bioessays (2012) 34:648–51. doi:10.1002/bies.201200005
355. Haase D, Roth O, Kalbe M, Schmiedeskamp G, Scharsack JP, Rosenstiel P, et al. Absence of major histocompatibility complex class II mediated immunity in pipefish, Syngnathus typhle: evidence from deep transcriptome sequencing. Biol Lett (2013) 9:20130044. doi:10.1098/rsbl.2013.0044
356. Grimholt U, Tsukamoto K, Azuma T, Leong J, Koop BF, Dijkstra JM. A comprehensive analysis of teleost MHC class I sequences. BMC Evol Biol (2015) 15:32. doi:10.1186/s12862-015-0309-1
357. Dijkstra JM, Grimholt U, Leong J, Koop BF, Hashimoto K. Comprehensive analysis of MHC class II genes in teleost fish genomes reveals dispensability of the peptide-loading DM system in a large part of vertebrates. BMC Evol Biol (2013) 13:260. doi:10.1186/1471-2148-13-260
358. Grimholt U. MHC and evolution in teleosts. Biology (Basel) (2016) 5:6–27. doi:10.3390/biology5010006
359. Dijkstra JM, Yamaguchi T, Grimholt U. Conservation of sequence motifs suggests that the nonclassical MHC class I lineages CD1/PROCR and UT were established before the emergence of tetrapod species. Immunogenetics (2017). doi:10.1007/s00251-017-1050-2
360. Chistiakov DA, Sobenin IA, Orekhov AN, Bobryshev YV. Myeloid dendritic cells: development, functions, and role in atherosclerotic inflammation. Immunobiology (2015) 220:833–44. doi:10.1016/j.imbio.2014.12.010
361. Haugland GT, Jordal AE, Wergeland HI. Characterization of small, mononuclear blood cells from salmon having high phagocytic capacity and ability to differentiate into dendritic like cells. PLoS One (2012) 7:e49260. doi:10.1371/journal.pone.0049260
362. Johansson P, Corripio-Miyar Y, Wang T, Collet B, Secombes CJ, Zou J. Characterisation and expression analysis of the rainbow trout (Oncorhynchus mykiss) homologue of the human dendritic cell marker CD208/lysosomal associated membrane protein 3. Dev Comp Immunol (2012) 37:402–13. doi:10.1016/j.dci.2012.02.012
363. Bassity E, Clark TG. Functional identification of dendritic cells in the teleost model, rainbow trout (Oncorhynchus mykiss). PLoS One (2012) 7:e33196. doi:10.1371/journal.pone.0033196
364. Granja AG, Leal E, Pignatelli J, Castro R, Abos B, Kato G, et al. Identification of Teleost skin CD8alpha+ dendritic-like cells, representing a potential common ancestor for mammalian cross-presenting dendritic cells. J Immunol (2015) 195:1825–37. doi:10.4049/jimmunol.1500322
365. Soleto I, Fischer U, Tafalla C, Granja AG. Identification of a potential common ancestor for mammalian cross-presenting dendritic cells in teleost respiratory surfaces. Front Immunol (2018) 9:59. doi:10.3389/fimmu.2018.00059
366. Shao T, Zhu LY, Nie L, Shi W, Dong WR, Xiang LX, et al. Characterization of surface phenotypic molecules of teleost dendritic cells. Dev Comp Immunol (2015) 49:38–43. doi:10.1016/j.dci.2014.11.010
367. Wcisel DJ, Yoder JA. The confounding complexity of innate immune receptors within and between teleost species. Fish Shellfish Immunol (2016) 53:24–34. doi:10.1016/j.fsi.2016.03.034
368. Rodriguez-Nunez I, Wcisel DJ, Litman GW, Yoder JA. Multigene families of immunoglobulin domain-containing innate immune receptors in zebrafish: deciphering the differences. Dev Comp Immunol (2014) 46:24–34. doi:10.1016/j.dci.2014.02.004
369. Montgomery BC, Cortes HD, Mewes-Ares J, Verheijen K, Stafford JL. Teleost IgSF immunoregulatory receptors. Dev Comp Immunol (2011) 35:1223–37. doi:10.1016/j.dci.2011.03.010
370. Zwozdesky MA, Fei C, Lillico DME, Stafford JL. Imaging flow cytometry and GST pulldown assays provide new insights into channel catfish leukocyte immune-type receptor-mediated phagocytic pathways. Dev Comp Immunol (2017) 67:126–38. doi:10.1016/j.dci.2016.10.011
371. Rombout JH, Van Der Tuin SJ, Yang G, Schopman N, Mroczek A, Hermsen T, et al. Expression of the polymeric immunoglobulin receptor (pIgR) in mucosal tissues of common carp (Cyprinus carpio L.). Fish Shellfish Immunol (2008) 24:620–8. doi:10.1016/j.fsi.2008.01.016
372. Zhang F, Liu D, Wang L, Li T, Chang Q, An L, et al. Characterization of IgM-binding protein: a pIgR-like molecule expressed by intestinal epithelial cells in the common carp (Cyprinus carpio L.). Vet Immunol Immunopathol (2015) 167:30–5. doi:10.1016/j.vetimm.2015.06.015
373. Yang S, Liu S, Qu B, Dong Y, Zhang S. Identification of sea bass pIgR shows its interaction with vitellogenin inducing antibody-like activities in HEK 293T cells. Fish Shellfish Immunol (2017) 63:394–404. doi:10.1016/j.fsi.2016.12.001
374. McKinney EC, Flajnik MF. IgM-mediated opsonization and cytotoxicity in the shark. J Leukoc Biol (1997) 61:141–6. doi:10.1002/jlb.61.2.141
375. Leiro J, Ortega M, Estevez J, Ubeira FM, Sanmartin ML. The role of opsonization by antibody and complement in in vitro phagocytosis of microsporidian parasites by turbot spleen cells. Vet Immunol Immunopathol (1996) 51:201–10. doi:10.1016/0165-2427(95)05509-6
376. Olivier G, Eaton CA, Campbell N. Interaction between Aeromonas salmonicida and peritoneal macrophages of brook trout (Salvelinus fontinalis). Vet Immunol Immunopathol (1986) 12:223–34. doi:10.1016/0165-2427(86)90126-1
377. Petit J, David L, Dirks R, Wiegertjes GF. Genomic and transcriptomic approaches to study immunology in cyprinids: what is next? Dev Comp Immunol (2017) 75:48–62. doi:10.1016/j.dci.2017.02.022
Keywords: teleost, monocyte, macrophages, antimicrobial, cytokine, respiratory burst, nitric oxide, nutrient depravation
Citation: Grayfer L, Kerimoglu B, Yaparla A, Hodgkinson JW, Xie J and Belosevic M (2018) Mechanisms of Fish Macrophage Antimicrobial Immunity. Front. Immunol. 9:1105. doi: 10.3389/fimmu.2018.01105
Received: 09 February 2018; Accepted: 02 May 2018;
Published: 28 May 2018
Edited by:
Brian Dixon, University of Waterloo, CanadaReviewed by:
Geert Wiegertjes, Wageningen University & Research, NetherlandsUwe Fischer, Friedrich Loeffler Institute Greifswald, Germany
Copyright: © 2018 Grayfer, Kerimoglu, Yaparla, Hodgkinson, Xie and Belosevic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miodrag Belosevic, mike.belosevic@ualberta.ca
 Leon Grayfer1
Leon Grayfer1 Jordan W. Hodgkinson
Jordan W. Hodgkinson Miodrag Belosevic
Miodrag Belosevic