- 1State Key Laboratory Breeding Base of Dao-di Herbs, National Resource Center for Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China
- 2Tianjin University of Traditional Chinese Medicine, Tianjin, China
- 3Department of Biological Sciences, National Sun Yat-sen University, Kaohsiung City, Taiwan
- 4Department of Biomedical Science and Environmental Biology, Kaohsiung Medical University, Kaohsiung City, Taiwan
Huang-lian (Coptis plants in China) are essential medicinal plants in China, C. chinensis var. chinensis and C. deltoidea have been domesticated and cultivated for 700 years. In this study, the genetic diversity patterns and biogeographical information of cultivated Huang-lian and their wild relatives Coptis species were assessed using three plastids DNA regions. A total of 186 individuals from twenty-seven populations representing two species of cultivated Huang-lian and four species of wild relatives were collected and analyzed. Twenty-four haplotypes of six species were identified when three plastid spacers were combined. Historical biogeography inference revealed multiple dispersal events in the groups of cultivated Huang-lian and C. omeiensis. This evidence can infer that large initial population size and interbreeding with co-existing wild relatives in expanding new planting areas might be the main reason for maintaining the high genetic diversity of cultivated Huang-lian. Nevertheless, the multimodal curve of mismatch analysis and positive or negative differed among species and populations by neutrality tests indicated some groups of cultivated Huang-lian experienced genetic bottlenecks. Phylogeny analysis (NJ, MP, BI) showed that cultivated Huang-lian and C. omeiensis were clustered into a monophyletic group while C. chinensis var. brevisepala was paraphyletic, having earlier divergence time from C. chinensis var. chinensis (7.6 Ma) than C. omeiensis. Parsimony network demonstrated that C. deltoidea had more shared haplotypes with C. omeiensis than C. chinensis var. chinensis, and other haplotypes of C. deltoidea and C. omeiensis had less mutation steps than that of C. chinensis var. chinensis and C. omeiensis. This evidence suggests that C. omeiensis has a closer relationship with cultivated Huang-lian and might be a potential wild relative to C. deltoidea. The results reported here provide the baseline data for preserving genetic resources of Huang-lian and also evaluating the genetic impacts of long-term cultivation on medicinal plants, which could be instructive to future cultivation projects of traditional Chinese medicinal plants.
Introduction
Wild plant populations are subjected to domestication and selective cultivation around 12,000–10,000 years ago in the Fertile Crescent and elsewhere (Xu, 2010). Cultivated plant species experienced long periods of artificial selection and isolation transferring to new cultivars and types to suit human needs (Doebley et al., 2006). In some cases, taxonomists have divided these wild and domesticated plants into two distinct species, such as Oryza sativa and O. rufipogon, Zea mays and Z. mexicana (Huang et al., 2012; Niazi et al., 2015). However, the domestication of the wild plant might have reduced genetic diversity (Allaby et al., 2019). Founder effect and artificial selection for high yielding or other good agronomic traits, have resulted in a narrow genetic material in cultivation and have usually been examined in cereal species (Doebley et al., 2006) such as maize (Tenaillon et al., 2004), wheat (Haudry et al., 2007), and rice (Londo et al., 2006). Therefore, it is essential to understand the genetic diversity of wild plant populations, as wild plant resources provide infinite potentials for superior breeding varieties to improve the genetic diversity of cultivated plants.
Cultivation of medicinal plants has been documented for more than 2000 years, which is shorter than crop domestication (12,000 years ago) (Xiao, 1991; Olsen and Gross, 2008). Medicinal plants contain 12.5% of the 422000 plant species documented worldwide playing a central role not only as traditional medicines systems but also as trade-in herbal medicinal products (Schippmann et al., 2002). In China, there are at least 6000 medicinal plants (Xiao, 1991; Huang, 2011). Of these, more than 600 plant species are commonly used in Chinese medicine, and approximately 300 species are cultivated with commercial use (Xiao, 1991; Bodeker et al., 1997). The demand for traditional Chinese medicine is increasing rapidly and is being cultivated on a large scale (Li et al., 2015). However, large-scale cultivation faces challenges of plant pests and diseases, heavy metal, agrochemicals residues and biodiversity conservation (Li et al., 2015).
The strength of the bottleneck effect of domesticated species is determined by the bottleneck’s duration and adequate population size of the bottlenecked population (Olsen and Gross, 2008). Several previous studies have pointed out that nearly all of the crops have experienced bottleneck effects and loss of genetic diversity relative to their wild species during long-term domestication (Hyten et al., 2006; Yamasaki et al., 2007; Kovach and Mccouch, 2008). The effect of domestication bottlenecks also revealed different patterns in perennial and annual cultivated crops (Miller and Gross, 2011). Compared with their wild population, perennial cultivated fruit crops preserve a greater proportion of genetic variation than annual cultivated fruit crops since juvenile phase length, mating system, mode of reproduction, geographic origins of cultivated individuals, widespread hybridization, and crop-wild gene flow (Miller and Gross, 2011). For these recently cultivated medicinal plants (about several decades), such as Scutellaria baicalensis and Gastrodia elata, experimental evidence indicates that a slight genetic bottleneck since there is not sufficient time for artificial selection and extensive seed exchange (Yuan et al., 2010; Chen et al., 2014). Compared with the recently cultivated perennial medicinal plants, the information about whether there is a stronger genetic bottleneck for the long-term domesticated perennial medicinal plant is poor. Thus, it is necessary to investigate the genetic diversity patterns of long-term cultivated medicinal plants and to elucidate the relationship with wild relatives.
Huang-lian (Rhizoma coptidis), is the rhizome of Coptis plants. Coptis is a pharmaceutically herbaceous perennial genus of Ranunculaceae (Yu et al., 2006). In China, there are six species of Coptis [C. chinensis var. chinensis, C. chinensis var. brevisepala W. T. Wang and P. K. Hsiao, C. omeiensis (Chen) C. Y. Cheng, C. deltoidea C. Y. Cheng and P. K. Hsiao, C. teeta Wallich, C. quinquesecta W. T. Wang] and wild populations of these species are restricted distribution, mainly distributed in Southern and Southwest of China and Himalayas (Tamura, 1995; Yu, 1999; Fu and Orbélia, 2001; Xiang et al., 2016, 2018). In Taiwan, there is one Coptis species (C. quinquefolia Miquel) with small population grow on restricted habitat in northern Taiwan (Tamura, 1995; Fu and Orbélia, 2001; Xiang et al., 2018). Huang-lian is well-known for its berberine-rich and broad antibacterial activities. Pharmacopeia-recorded Huang-lian derived from three Coptis species: C. chinensis var. chinensis, C. deltoidea and C. teeta (China, 2015), which is called official Huang-lian. The other Coptis species are used as alternative Huang-lian in folk. C. chinensis var. chinensis, C. omeiensis and C. teeta are diploid (2n = 2x = 18), whereas C. deltoidei is auto triploid (2n = 3x = 27) (Pandit and Babu, 1993; Xiang et al., 2010; Huang et al., 2013). C. chinensis var. chinensis and C. omeiensis are propagated by seeds and no stolons (Song et al., 2010). C. deltoidei is propagated by stolons while is cannot form seeds (Li et al., 2010). C. teeta is propagated by seeds and stolons (Selvam and India, 2012). In China, the wild resources of these species are almost endangered (He et al., 2014; Zhong et al., 2018). C. omeiensis has not been artificially cultivated, and the source all originated from wild populations in China (Huang, 2012). C. omeiensis was listed as a national second-class endangered plant in China (Huang, 2012). C. teeta had been listed in the International Union for Conservation of Nature’s Red List (IUCN Red List) as Endangered (Saha et al., 2015). C. teeta has been cultivated and domesticated on a small scale (Mukherjee and Chakraborty, 2019). Compared with wild C. teeta, the cultivated C. teeta has been observed obvious morphological variations (Mukherjee and Chakraborty, 2019). C. chinensis var. chinensis and C. deltoidea have been artificially cultivated since the Yuan and Ming Dynasty (Xiong et al., 2011). Currently, C. chinensis var. chinens is cultivated in Chongqing, Hubei and Sichuan provinces (Lv et al., 2016). C. deltoidea is cultivated on both sides of Emeishan in Sichuan province (Xiong et al., 2011). C. chinensis var. chinens is a common and high-yielding material of Huang-lian. The information on the distribution, ploidy level, propagation and present cultivation status of six Coptis species are listed in Supplementary Table S1.
Huang-lian was first recorded in the earliest monograph on Chinese material medical, Sheng Nong’s Herbal Classic during the Eastern Han Dynasty (25–220 AD) (Yang, 1998; Wu, 2005). Huang-lian has been used by Chinese medicinal physicians for over 2000 years and has been domesticated and cultivated for 700 years in China (Tan et al., 2016). Therefore, it is an ideal model for exploring the effects of long-term domesticated and cultivated medicinal plants on genetic diversity patterns. Cultivated Huang-lian has abundant wild relative species resources. C. teeta is an endemic endangered plant of Eastern Himalaya. It is only reported from Indian Territory of Arunachal Pradesh, Burma, northwest Yunnan and Tibet provinces of China (Payum, 2017; Wang et al., 2019). The distribution of C. teeta does not overlap with C. chinensis var. chinensis and C. deltoidea. Wild Coptis of C. omeiensis is mainly distributed on Emeishan in Sichuan province, overlapping with the distribution of C. deltoidea. C. chinensis var. brevisepala is a wild plant distributed in the Southern Anhui mountainous areas, partially overlapping with the distribution of C. chinensis var. chinensis (Fu and Orbélia, 2001; Zhang and Zhang, 2005; Wang and Peng, 2008). C. quinquesecta is a critically endangered wild species native in southeastern Yunnan Province (Zhang et al., 2018), and has not found according to herbaria records.
Based on plastid and ITS DNA regions evidence, C. quinquefolia in Taiwan has a relatively distant relationship with the other six Coptis species in China (Xiang et al., 2016). Furthermore, C. chinensis var. chinensis and C. chinensis var. brevisepala are not clustered in a group. Therefore, Xiang et al. suggest that C. chinensis var. brevisepala should be regarded as a species rank (Xiang et al., 2016). According to metabolic and molecular genetic information, C. omeiensis and C. deltoidea had the closest genetic relationship among Coptis plants (He et al., 2014; Xiang et al., 2016; Chen et al., 2017; Zhong et al., 2018). There are only a few molecular phylogeny studies of genus Coptis have been reported so far (He et al., 2014; Xiang et al., 2016; Chen et al., 2017; Zhong et al., 2018). Only some researches have been done to gain information about the genetic diversity of Coptis species using molecular methods such as Inter-simple sequence repeats (ISSR) (Chen et al., 2006; Shi et al., 2008). However, less attention was paid to the comprehensive genetic evaluation of Coptis species with medicinal effects. There is urgent to explore the genetic information between long-term domesticated Huang-lian and their wild populations or wild relative species.
Nowadays, the assessment of genetic diversity is mainly based on DNA-based methods, which can be classified into fragment analysis-based methods, hybridization array-based methods, and sequencing-based methods (Loera-Sánchez et al., 2019). The use of such methods to assess the genetic diversity will require careful evaluation and balance of the biological features of the plant species, and the strengths and limitations of the method used, in order to optimally exploit suitable techniques for study genetic diversity. Among them, using amplicon sequencing to assess the genetic diversity is a low-cost and complexity reduction alternative method since only standard PCR reagents are required. It has been applied to assess the genetic diversity information and a monitoring tool for mixture breeding in various plant species (Loera-Sánchez et al., 2019). Therefore, this study employs DNA sequencing technologies to assess the phylogenetic relationships and genetic diversity of Huang-lian and wild relatives, which can help to establish and monitoring genetic resource conservation and can also support the species registration.
Phylogeography integrates genealogical and geographical information to understand the demographic and historical processes that shaped the evolution of populations or closely related species (Avise, 2000; Kuchta and Meyer, 2001). Phylogeographical approaches are well suited for understanding the domestication processes and history because domesticated species usually include many different degrees of divergence in the evolutionary lineages resulting from the several population recent differentiation processes (Aguirre-Dugua and González-Rodríguez, 2016). Here, we applied three plastids and one nuclear (ITS) DNA regions to investigate the genetic variability and phylogenetic relationships of six Coptis species and analyzed the biogeographical information to clarify the evolutionary process. The objectives of this study are (1) to evaluate the genetic diversity pattern of cultivated Huang-lian; (2) to elucidate the phylogenetic and phylogeographical relationships among wild relatives Coptis species and cultivated Huang-lian
Materials and Methods
Plant Material Sampling
A total of 248 individuals from twenty-seven populations were collected and analyzed in this study (Table 1), including C. chinensis var. chinensis (Ccc: 38 individuals from 5 cultivated populations), C. deltoidea (Cd: 46 individuals from 5 cultivated populations), C. chinensis var. brevisepala (Ccb: 51 individuals from 4 wild populations), C. teeta (Ct: 38 individuals from 5 wild populations), C. omeiensis (Co: 65 individuals from 7 wild populations) and C. quinquefolia (Cq: 10 individuals from 1 wild population). This study contained most of the Coptis species in Taiwan and China but did not include C. quinquesecta, because the species is a critically endangered medicinal plant and was not found in the field. Fresh leaves of Coptis species were collected and dried with silica gel for DNA extraction. Voucher specimens were deposited in herbaria of Institute of Chinese Materia Medica (CMMI), China Academy of Chinese Medical Sciences. The precise geographic location of each sampled population was determined using a Garmin GPS unit (Table 1). The abbreviation naming rules are follows: species abbreviation + (population abbreviation) + W (wild) or C (cultivated).
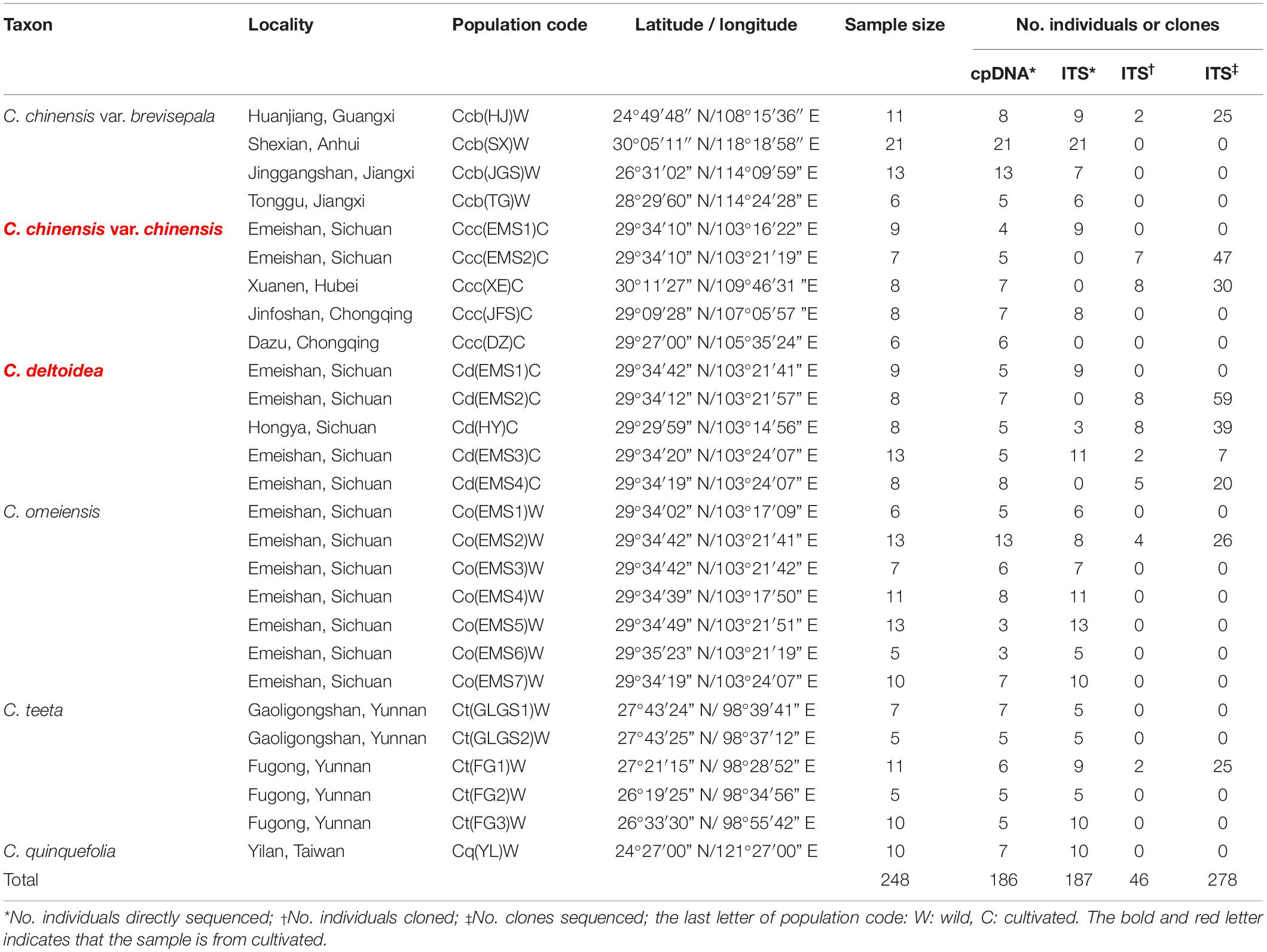
Table 1. Details of sample locations, sample sizes and the number of individuals and clones sequenced of cpDNA and ITS.
DNA Extraction, PCR Amplification, and Sequencing
Genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) protocol (Doyle and Doyle, 1987). Four DNA-barcoding regions recommended by China Plant BOL Group, i.e., chloroplast trnH-psbA, rbcL, matK, and nuclear ribosomal (nr) ITS were amplified and sequenced (Li et al., 2011). The primers suggested for rbcL (1F-724R) and matK (3F-1R) by the CBOL Plant Working Group (2009), trnH-psbA by Sang et al. (1997), and ITS (ITS5-ITS4) by White et al. (Innis et al., 1990) were used in this study. DNA amplification was performed in a TC-512 thermocycler (Techne, United Kingdom), programmed for a primary denaturing step at 94°C for 240 s, followed by 35 cycles of 45 s at 94°C, annealing at a specific temperature for 30 s and extension 90 s at 72°C, with a final extension of 10 min at 72°C. Reactions were carried out in a volume of 20 μL containing 2.0 mmol/L MgCl2, 0.5 μmol/L dNTP, 10 × buffer, 2.5 μmol/L primer, 1.0 U Ex-Taq DNA polymerase and 20 ng DNA template.
Sequencing reactions were conducted with the forward or reverse primers of the amplification reaction using the DYEnamic ET Terminator Kit (Amersham Pharmacia Biotech) following the manufacturer’s protocol. Sequencing was performed on an ABI 377XL automated DNA sequencer after the products of the direct sequencing reactions were purified through precipitation with 95% ethanol and 3 M sodium acetate (pH 5.2).
For those PCR products of ITS failed to directly sequence, some of them were cloned into the pGEM-T easy-cloning vector (TaKaRa) to separate multicopy or allele within the individual. Two to eight clones each PCR product were selected and reamplified. The direct cycle sequencing reactions were performed using a Thermo Sequenase cycle sequencing kit (rpn2438 from Amersham Pharmacia Biotech) with IRD700- and IRD800-labeled primers (MWG Biotech, Ebersberg, Germany). Sequencing was performed on a LICOR L4200S-2 automated sequencer.
Data Analysis
All the sequences were edited and assembled by using CONTIGEXPRESS (VECTOR NTI ADVANCE version 10.3.1, Invitrogen) and BIOEDIT 7.0.9 to assisted with manual sequence correction. Refer to the methods of Caicedo and Schaal, nucleotide insertion and deletion (indel) were coded as substitutions (Caicedo and Schaal, 2004). Sequence alignments for each region were carried out using CLUSTAL_X version 2.0 (Larkin et al., 2007) and then checked by eye in BIOEDIT. Accessions from each species were incorporated into one or more private sequences to checking the shared alleles among species by using DAMBE (Xia and Xie, 2001). Before conducting all analyses, the incongruence length difference (ILD) test (Farris et al., 1994) was conducted to evaluate congruency between data from the three cpDNA regions implemented in PAUP∗ 4.0b10 (Swofford, 2002).
Genetic diversity information was evaluated based on the number of segregating sites (S), the number of haplotypes (h), haplotype diversity (Hd) and nucleotide diversity (π) (Nei, 1987) using DNASP 5.10.1 (Librado and Rozas, 2009). Tests of neutrality and determination of the associated significance were carried out based on Tajima’s D statistic (Tajima, 1989) and Fu and Li’s D∗ statistic (Fu and Li, 1993) using DNASP 5.10.1 (Librado and Rozas, 2009). Pairwise FST-values among populations and species were also estimated hierarchically in program DNASP 5.10.1 (Librado and Rozas, 2009). The genetic structure was evaluated to estimate the distinction among species in Coptis by AMOVA analysis with ARLEQUIN version 3.1 (Excoffier et al., 2005), partitioning the genetic diversity into three levels: among species, among-population within species and within-population. Significance tests were conducted using 10,000 permutations. A statistical parsimony haplotype network estimated the genetic relationships among haplotypes by using TCS 1.21 (Clement and Crandall, 2000).
Phylogenetic Analyses
Phylogenetic relationships of haplotypes were inferred using neighbor-joining (NJ), Maximum parsimony (MP) and Bayesian Inference (BI) methods at the same time. NJ phylogenetic tree was constructed by using MEGA X software under the Tamura 3-parameter model (Kumar et al., 2018). MP phylogenetic tree analyses were carried out in PHYLIP Version 3.695 (Felsenstein, 1993), calculated by the modules SEQBOOT, DNAPARS, and CONSENSE. Phylogenetic trees were generated by maximum parsimony (MP) methods in the DNAPARS program, SEQBOOT program (1000 replicates) for generating bootstrap values and CONSENSE program to find a consensus tree. For Bayesian analysis, we chose Bayesian methods implemented in MRBAYES 3.1.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) with the best-fitting GTR+I+G model selected by MODELTEST 3.7 (Posada, 1998) using the Akaike information criterion (Akaike, 1974). Four chains, each with a different starting seed, were run for two million generations; each analysis was repeated twice. Trees were subsequently sampled every 100 generations. Stationarity was reached after approximately 400,000 generations, therefore, the first 4000 trees were discarded as burn-in. The general criteria for determining stationarity has been reached was when the value of the likelihood of the sample points reached a stable equilibrium (Huelsenbeck and Ronquist, 2001). The value was the standard deviation of the split frequencies was less than 0.01, with the potential scale reduction factors for each parameter was close to 1. Bayesian posterior probabilities were estimated as the proportion of trees containing each node relative to the total tree number sampled during runs.
Divergence time estimation among and within species was carried on BEAST 1.7.2 (Drummond and Rambaut, 2007). As a taxon-specific substitution rate had not been previously calibrated for non-coding cpDNA regions of Coptis species, we assumed a substitution rate of 1.52 × 10–9 substitutions per neutral site per year (s/s/y) and set 1.0 × 10–9 and 3.0 × 10–9 as the lower and the upper limit for nucleotide substitution rate, respectively (Wolfe et al., 1987). Three independent Markov chain Monte Carlo (MCMC) pre-runs of ten million generations were performed. Genealogies were sampled every 1000 generations with the first 10% discarded as burn-in. Statistics of the output values were summarized using Tracer 1.5 (Rambaut and Drummond, 2009), and both log and tree files were combined using LOGCOMBINER 1.6.1 (Drummond and Rambaut, 2007). We applied TREEANNOTATOR 1.6.1 (Drummond and Rambaut, 2007) and FIGTREE 1.4 (Rambaut, 2012) to summarize and display the sampled trees, respectively.
Bayesian skyline plot (BSP) was created to infer the historical demography of all species using by running BSP analysis using the software BEAST 1.7.2. A GTR+I+G model was selected with an assuming nucleotide substitution rate of 1.52 × 10–9 s/s/y. Three independent Markov chains were run for 2.0 × 107 generations and were sampled every 1000 generations discarding the first 10% as burn-in. Mismatch analysis was performed to evaluate the range expansions of the Coptis species under the sudden expansion model and the spatial expansion model using Arlequin ver 3.5 (Excoffier and Lischer, 2010). The sum of squared deviations (SSD) was used as a statistical method to test the validity of the expansion model. P-values were then calculated as the proportion of simulations generating an SSD more substantial than the observed SSD. The raggedness index (HRag) and its significance for observed distributions were used to compute the smoothness of the mismatch distributions (Harpending, 1994). Low raggedness implied recently non-stationary, expanding populations.
Phylogeographical Inference Using RASP
To reveal the biogeographical history, the origin and mechanism (vicariance or dispersal) of the geographical distribution of Coptis, statistical dispersal-vicariance analysis (S-DIVA) and Bayesian binary MCMC (BBM) analysis performed using RASP (Reconstruct Ancestral State in Phylogenies) (Yu et al., 2015). The geographical distribution of the species was categorized into seven areas: (A) Yunnan province (5 populations of C. teeta), China; (B) Sichuan province (all the populations of C. deltoidea and C. omeiensis, and two populations of C. chinensis var. chinensis), China; (C) Taiwan Island (unique population of C. quinquefolia), China; (D) Anhui province (only Ccb (SX)W), China; (E) Guangxi province (only Ccb(HJ)W), China; (F) Chongqing municipality and Hubei province (three populations of C. chinensis var. chinensis), China; and (G) Jiangxi province (Ccb(JGS)W and Ccb(TG)W), China. The tree topologies generated by MRBAYES and BEAST were congruent in the major clades; therefore, 20,000 trees from the BEAST MCMC outputs were used, and the BEAST annotated tree was set as the final condensed tree. The number of maximum areas at each node allowed in ancestral distributions was kept as three. The other parameters were automatically optimized.
Results
Sequence Characteristics of Chloroplast DNA and ITS
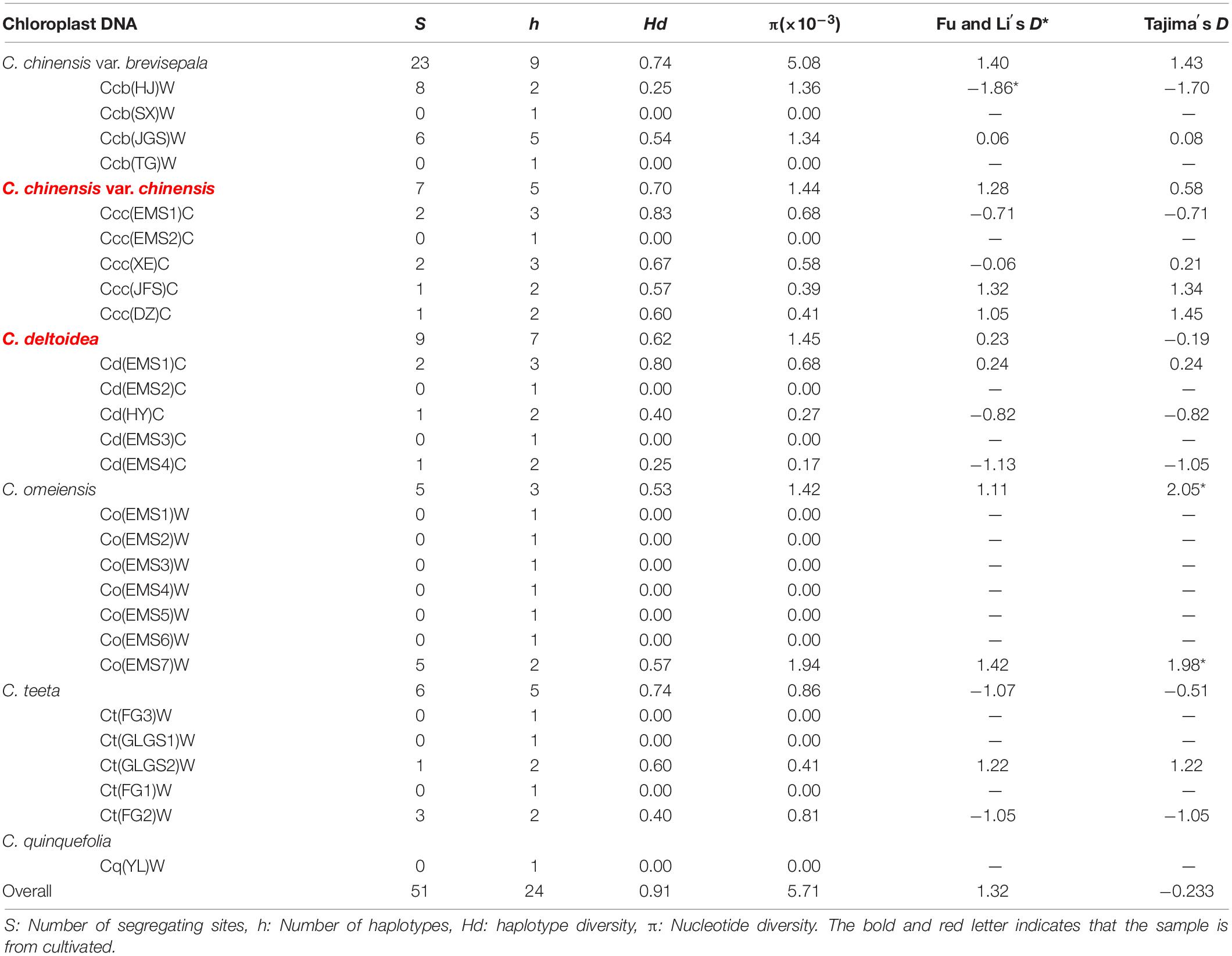
A total of 186 individuals representing twenty-seven populations of six species or variation of Coptis were successfully amplified and sequenced using chloroplast trnH-psbA, rbcL and matK. The newly generated sequences in this study were submitted to GenBank (Supplementary Table S2). Sequence lengths after alignment were 373, 669 and 727 bp for trnH-psbA, rbcL and matK, respectively. The matK region was the most variable (35 polymorphisms detected in 727 aligned positions; 4.81%), followed by trnH-psbA (17/373, 4.56%) and rbcL (9/669, 1.35%). The matK and trnH-psbA region had 55 indels of 1−5 bp and 10 indels of 1−70 bp, respectively. The rbcL matrix had no indels. The results of the incongruence length difference (ILD) test suggested a high degree of homogeneity among the three cpDNA regions (P > 0.5), which therefore were combined and formed twenty-four haplotypes (Hap1-24) based on 51 segregating sites (Tables 1, 2 and Supplementary Table S2).
ITS was difficult to sequence directly by PCR in some populations of Coptis species, especially in C. chinensis var. chinensis and C. deltoidea. Sequencing by clone was performed in most populations of the two species and a few populations of the remaining species. The sequences by PCR and by clone presented 174 haplotypes (H1-174) and were submitted to GenBank (Table 1 and Supplementary Table S2), which showed significant intragenomic variability (average of 3.78 polymorphisms per individual). The results suggest that ITS may be incomplete concerted evolution in Coptis and was not suitable for phylogenetic analyses and phylogeographical inference. Therefore, only chloroplast DNA sequences were used in the following analyses.
Genetic Diversity and Differentiation of Coptis Species
Genetic variability of cpDNA was estimated by the number of haplotypes, haplotype diversity and nucleotide diversity in Coptis species (Table 2). Haplotype Hap1, with the highest haplotype frequency (19.4%), was shared between cultivated Huang-lian, C. chinensis var. chinensis and C. deltoidea, and wild C. omeiensis (Figure 1 and Supplementary Table S2). C. deltoidea shared four haplotypes (Hap1, Hap4-6) with C. chinensis var. chinensis and two haplotypes (Hap1, Hap3) with C. omeiensis, respectively. Only two or one private haplotype were detected in the three species (C. deltoidea: Hap8-9, C. chinensis var. chinensis: Hap15, and C. omeiensis: Hap2). Among the remaining three species, no haplotypes were shared, nine (Hap7, 16-23), five (Hap10-14) and one (Hap24) private haplotypes were fixed in C. chinensis var. brevisepala, C. teeta and C. quinquefolia, respectively. The proportion of the population with unique haplotype in cultivated Huang-lian (C. chinensis var. chinensis: 1/5; C. deltoidea: 2/7) was lower than that in wild Coptis species (C. omeiensis: 1/4; C. chinensis var. brevisepala: 9/9; C. teeta: 5/5; C. quinquefolia: 1/1) (Figure 1).
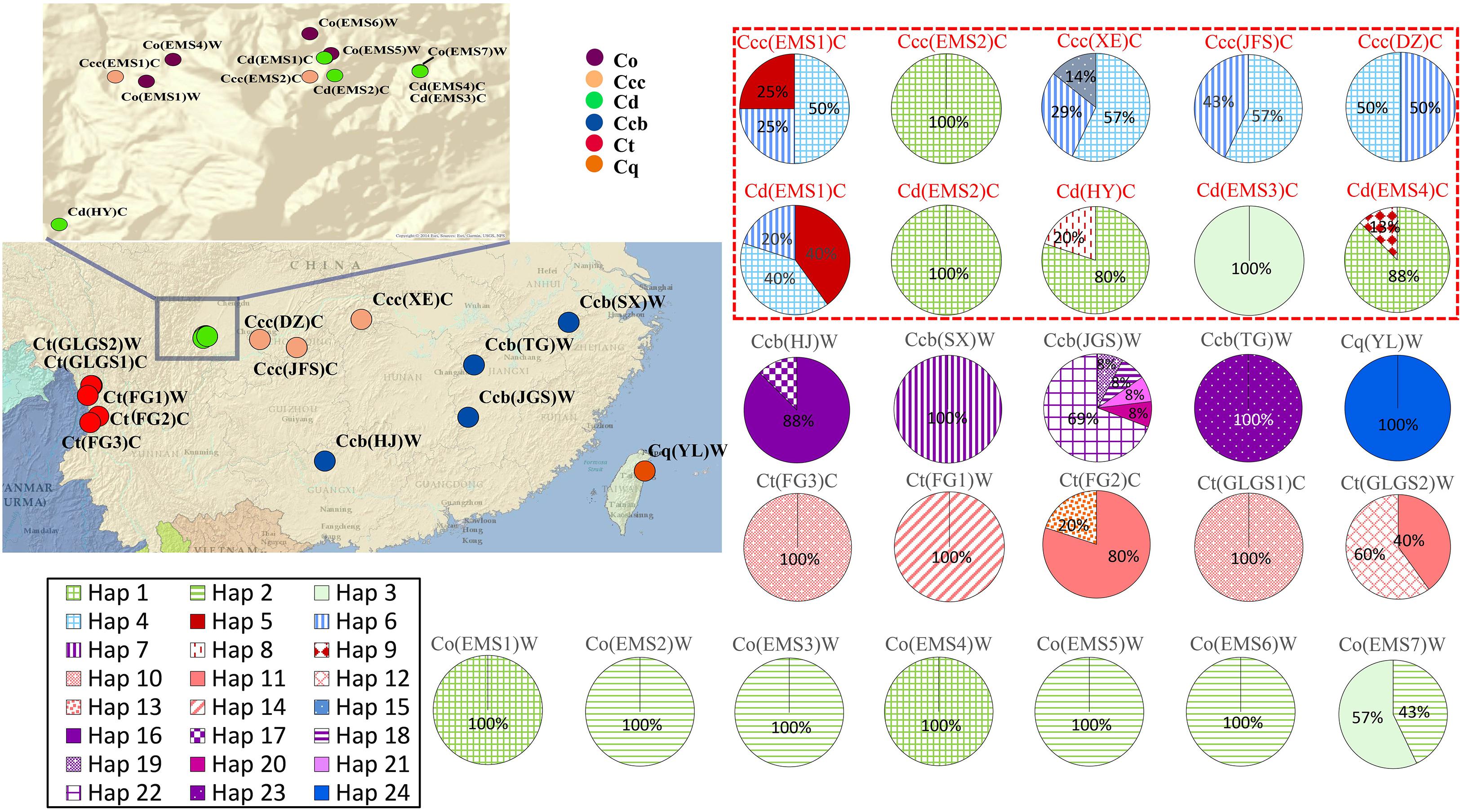
Figure 1. Geographic distribution and haplotype network of cpDNA haplotypes in populations of C. chinensis var. brevisepala (Ccb), C. chinensis var. chinensis (Ccc), C. deltoidea (Cd), C. omeiensis (Co), C. teeta (Ct), and C. quinquefolia (Cq). Pie charts show haplotype proportions for site locations. See Table 1 for abbreviation codes. The red letter indicates that the sample is from cultivated.
In the haplotype data of ITS, haplotype H2 with the highest haplotype frequency (16.0%), was shared between cultivated Huang-lian of C. deltoidei and wild C. omeiensis (Supplementary Tables S2, S3 and Figure 2). C. deltoidea shared eleven haplotypes (H1, H14, H19, H21-23, H50, H58, H65-66, H79) with C. chinensis var. chinensis and one haplotype (H2) with C. omeiensis, respectively. C. chinensis var. chinensis shared one haplotype (H30) with C. chinensis var. brevisepala (Figure 2 and Supplementary Tables S2, S3). C. teeta did not share any genotypes with other species. C. deltoidei had the largest number of private genotypes (Figure 2 and Supplementary Tables S2, S3).
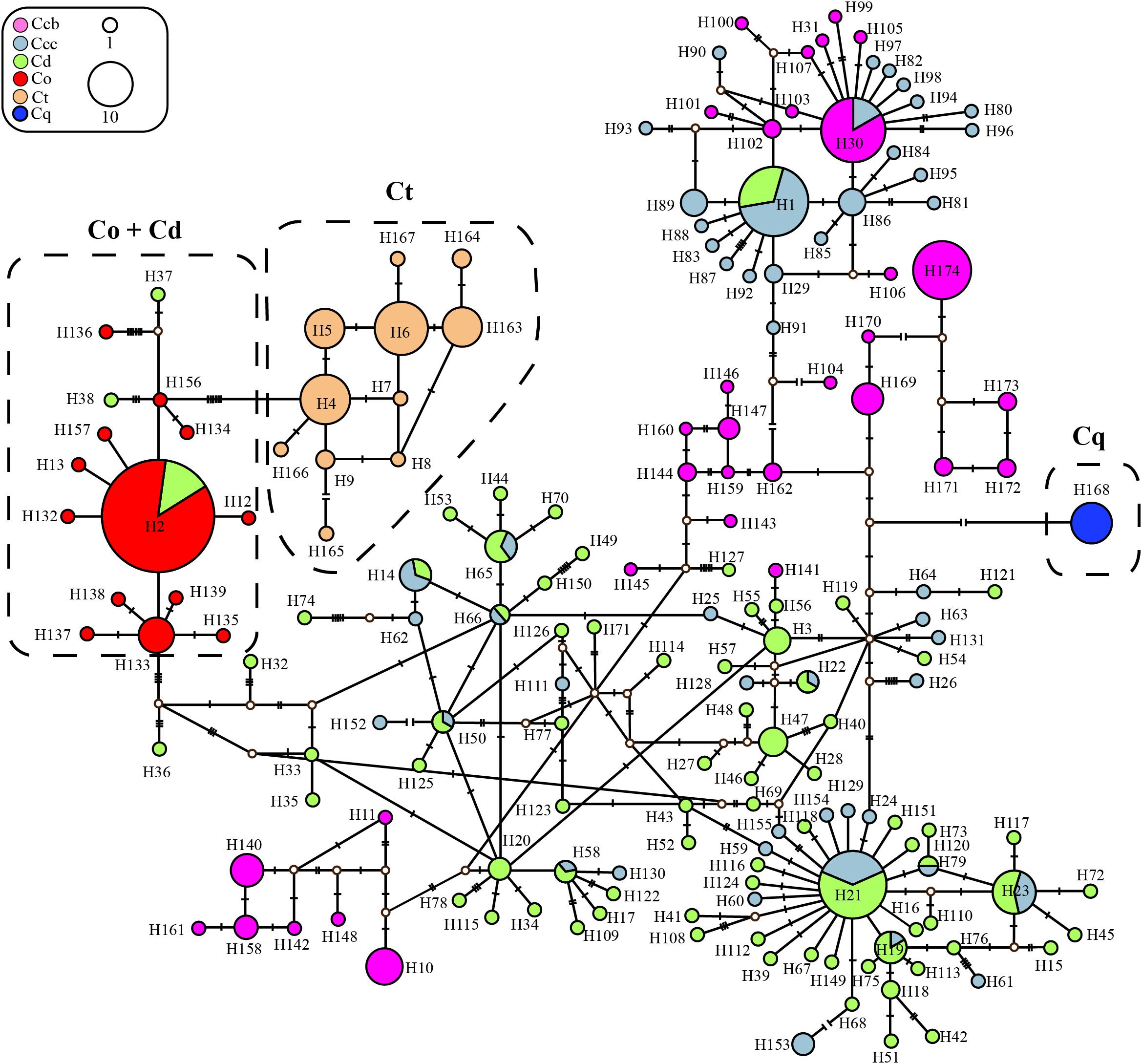
Figure 2. Parsimony network for all ITS haplotypes of C. chinensis var. brevisepala (Ccb), C. chinensis var. chinensis (Ccc), C. deltoidea (Cd), C. omeiensis (Co), C. teeta (Ct), and C. quinquefolia (Cq) species based on cpDNA data. The relative size of each circle corresponds to proportional to haplotype frequencies. The small hollow dots indicate hypothetical missing haplotypes. The network shown in different colors correspond to each species.
Total haplotype diversity (Hd) and nucleotide diversity (π) in all species of Coptis were 0.91 and 0.00571, respectively (Table 2). Haplotype diversity and nucleotide diversity of cultivated Huang-lian (Ccc: 0.70, 0.00144; Cd: 0.62, 0.00145) was higher than that of C. omeiensis (0.53; 0.00142) and lower than that of C. chinensis var. brevisepala (0.74; 0.00508). However, the haplotype diversity (0.74) and nucleotide diversity (0.00086) of C. teeta were inconsistent. The former was higher while the latter was lower than those of cultivated Huang-lian. In summary, the genetic diversity of cultivated Huang-lian has not reduced compared with three wild Coptis species. However, the genetic diversity within populations of cultivated Huang-lian significantly raised. The top two Hd were observed in two cultivated populations Ccc(EMS1)C (0.83) and Cd(EMS1)C (0.80) in order. The Hd of C. chinensis var. chinensis (>0.57 except Ccc(EMS2)C) was higher than that of its wild variety, C. chinensis var. brevisepala (<0.54). The Hd of C. deltoidea (>0.25 except Cd(EMS2)C and Cd(EMS3)C) was higher than that of sympatric wild Coptis species, C. omeiensis ( = 0 except Co(EMS7)W). These results indicated human activities have increased a potential gene flows among cultivated populations during domestication.
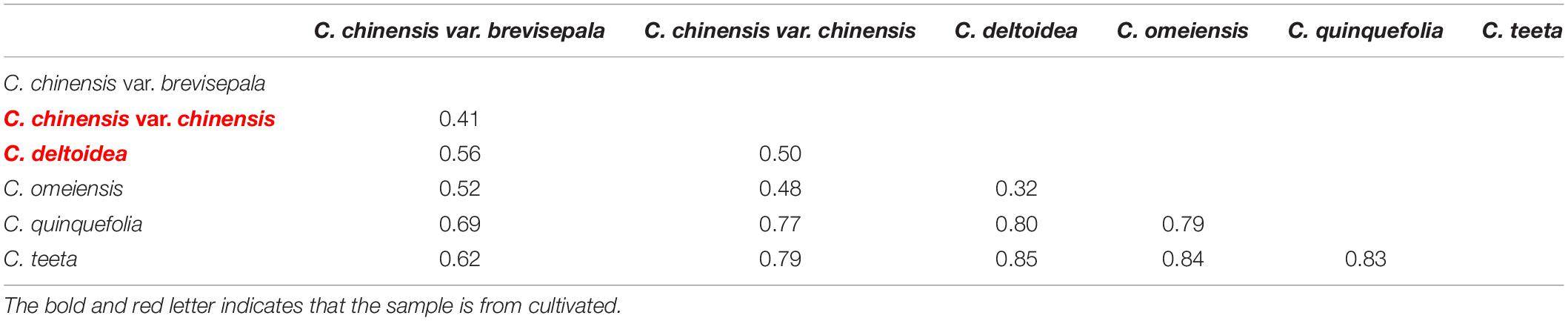
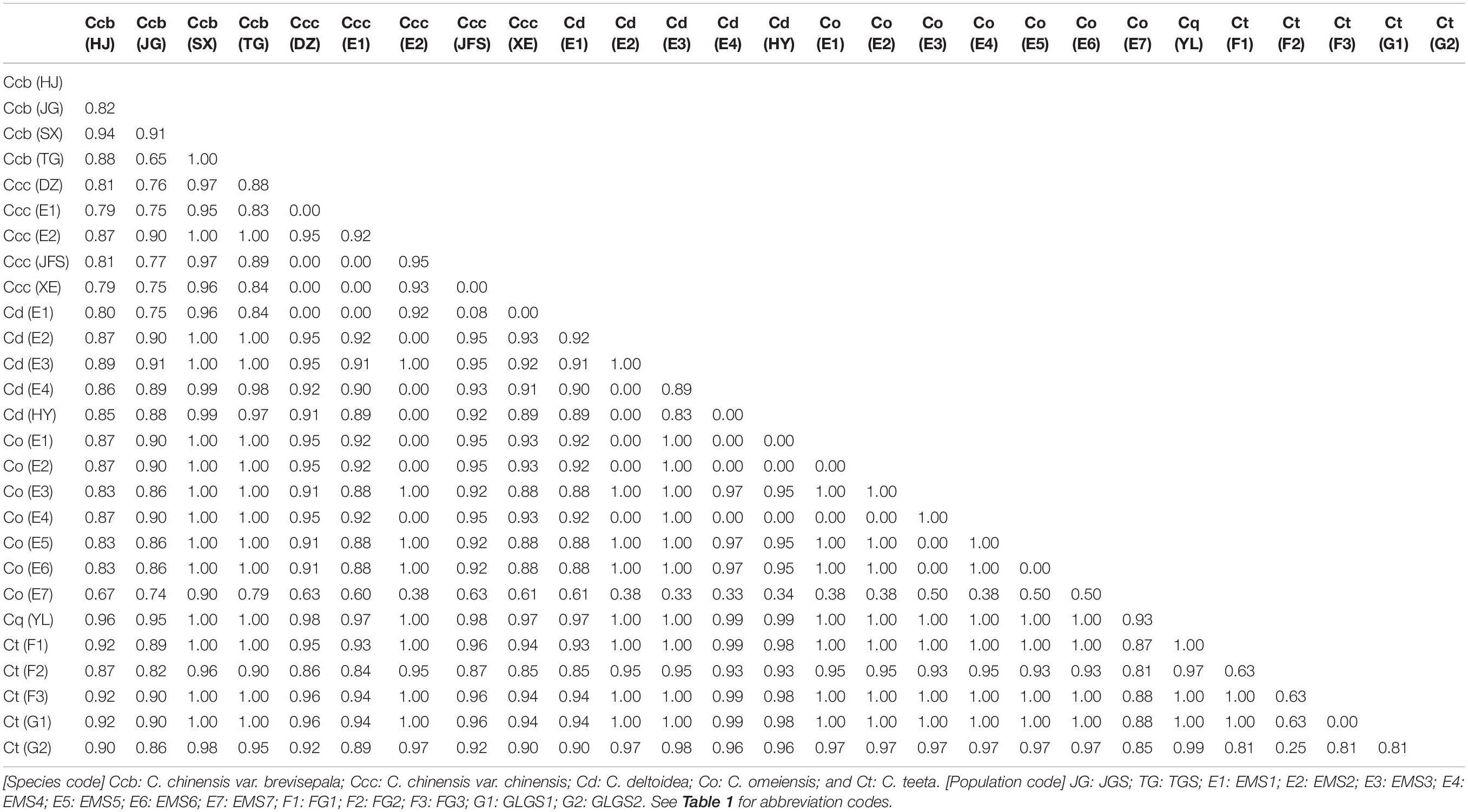
Genetic differentiation between Coptis species was estimated based on FST (Table 3). The genetic differentiation between C. deltoidea and C. omeiensis (FST = 0.32) was the least in those between C. deltoidea and other species of Coptis. The genetic differentiation between C. chinensis var. chinensis and C. chinensis var. brevisepala (FST = 0.41) was the least in those between C. chinensis var. chinensis and other species of Coptis, but was higher than that between C. deltoidea and C. omeiensis (FST = 0.32). C. quinquefolia (FST>0.69) and C. teeta (FST>0.62) have significant divergence from other species of Coptis (Table 3). Among intraspecies populations, cultivated Huang-lian showed lower genetic differentiation and a certain degree of homogenization (Table 4). Only one population (Ccc (E2)) of C. chinensis var. chinensis (FST>0.92) and two populations (Cd (E1) and Cd (E3)) of C. deltoidea (FST>0.83) were significantly divergent from other intraspecies populations, while the remaining populations of the two species had no differentiation each other (FST = 0). Wild Coptis species with high haplotype diversity presented relatively high divergence among intraspecies populations, FST-values ranging from 0.65 to 1.00 for the populations of C. chinensis var. brevisepala. FST-values ranging from 0.25 to 1.00 for the populations of C. teeta, except for the differentiation between Ct (F3) and Ct (G1) populations (FST = 0) (Table 4). FST-values ranging from 0.38 to 1.00 for the populations of C. omeiensis, except for the differentiation between Co (E1) and Co (E2), Co (E1) and Co (E4), Co (E2) and Co (E4), Co (E3) and Co (E5), Co (E3) and Co (E6), Co (E5) and Co (E6) populations (FST = 0) (Table 4).
Hierarchical AMOVA based on cpDNA data revealed that 5.49% of the total variance was distributed within populations (ΦST = 0.95, P < 0.05), and a considerable amount of variation originated from differentiations among populations within species (38.15%, ΦSC = 0.87, P < 0.05) and over a half of all variance distinguished different species from each other (56.36%, ΦCT = 0.56, P < 0.05) (Table 5). For each species, C. chinensis var. chinensis had the lowest genetic variation among populations (ΦST = 0.75) and C. chinensis var. brevisepala had the highest genetic variation among populations (ΦST = 0.92).
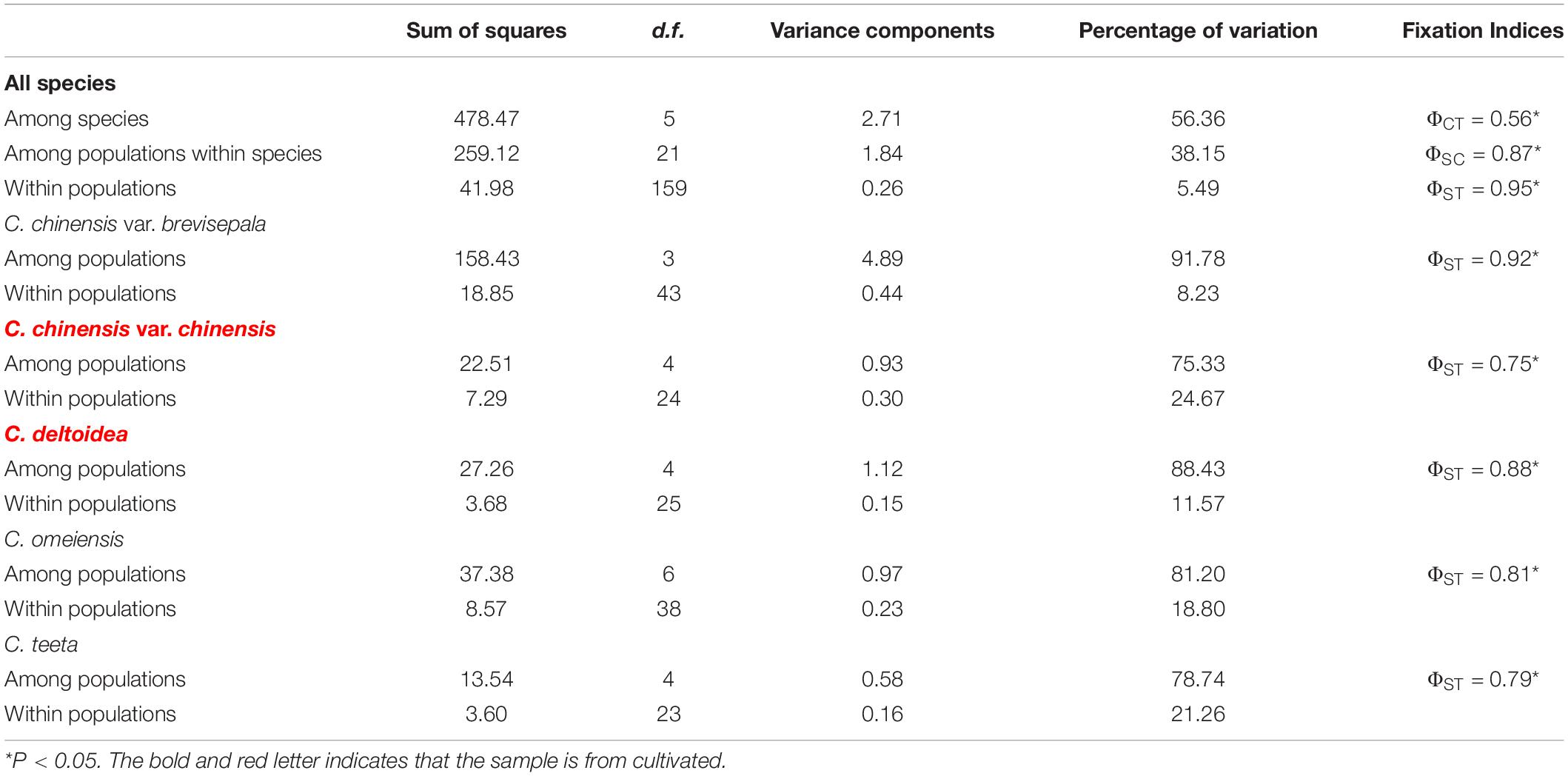
Table 5. Results of different hierarchical types analysis of molecular variance (AMOVA) of chloroplast DNA sequence data.
Phylogenetic Relationships of cpDNA Haplotypes and Divergence Time Estimates
Three phylogeny analysis methods (NJ, MP, BI) generated congruent tree topologies in terms of the major clades (Figure 3). Overall, the 24 cpDNA haplotypes were clustered into four groups corresponding to individual species. All haplotypes of cultivated Huang-lian and C. omeiensis (Hap1-6, 8-9, 15) were included in a monophyletic group with 37% (NJ) or 33% (BI) bootstrap support values, of which paraphyletic clade was the group contained all haplotypes of C. chinensis var. brevisepala except Hap16. Haplotypes of C. teeta (Hap 10-14) formed a monophyletic group with firm support. The haplotype of C. quinquefolia was distinctively apart. Similar results were depicted by the statistical parsimony network (Figure 4). Haplotypes of cultivated Huang-lian and C. omeiensis connected to form a network. Haplotypes of C. chinensis var. brevisepala formed a relatively independent clade network connected to Hap6 of cultivated Huang-lian except for Hap16 (Corresponding to the H10, H11, H140, H142, H148, H161 of ITS haplotypes). Haplotypes of C. teeta and C. quinquefolia grouped into two significant divergent clades with relatively far from the other four species by more than four mutational steps. This similar grouping patterns can also be obtained from statistical parsimony network results of ITS data (Figure 2). Haplotypes of C. omeiensis formed an intermediate network and shared H2 of ITS haplotypes with cultivated C. deltoidea and two haplotypes (H37 and H38 of ITS haplotypes) occupying tips belong to cultivated C. deltoidea (Figure 2).
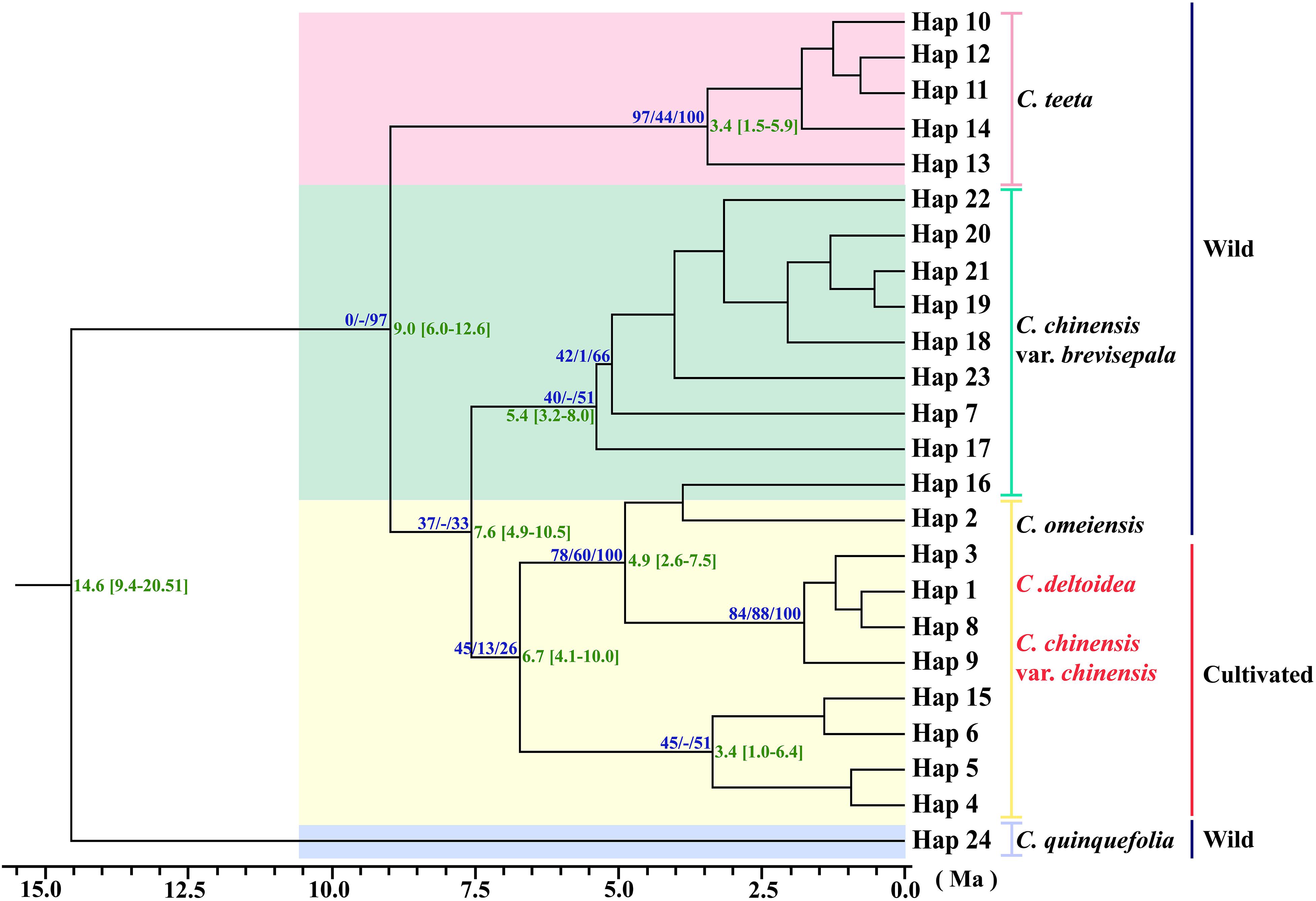
Figure 3. Phylogenetic relationships analysis using Maximum Parsimony (MP) methods based on cpDNA data. The numbers above the branches are bootstrap support values of Neighbor-Joining and Maximum parsimony tree, and Bayesian posterior probabilities (PPs) of 100% of BI tree (NJ/MP/BI) between major lineages. Time scale (with 95% HPD intervals in square brackets) are shown to the right of each node with green color. The red letter indicates that the sample is from cultivated.
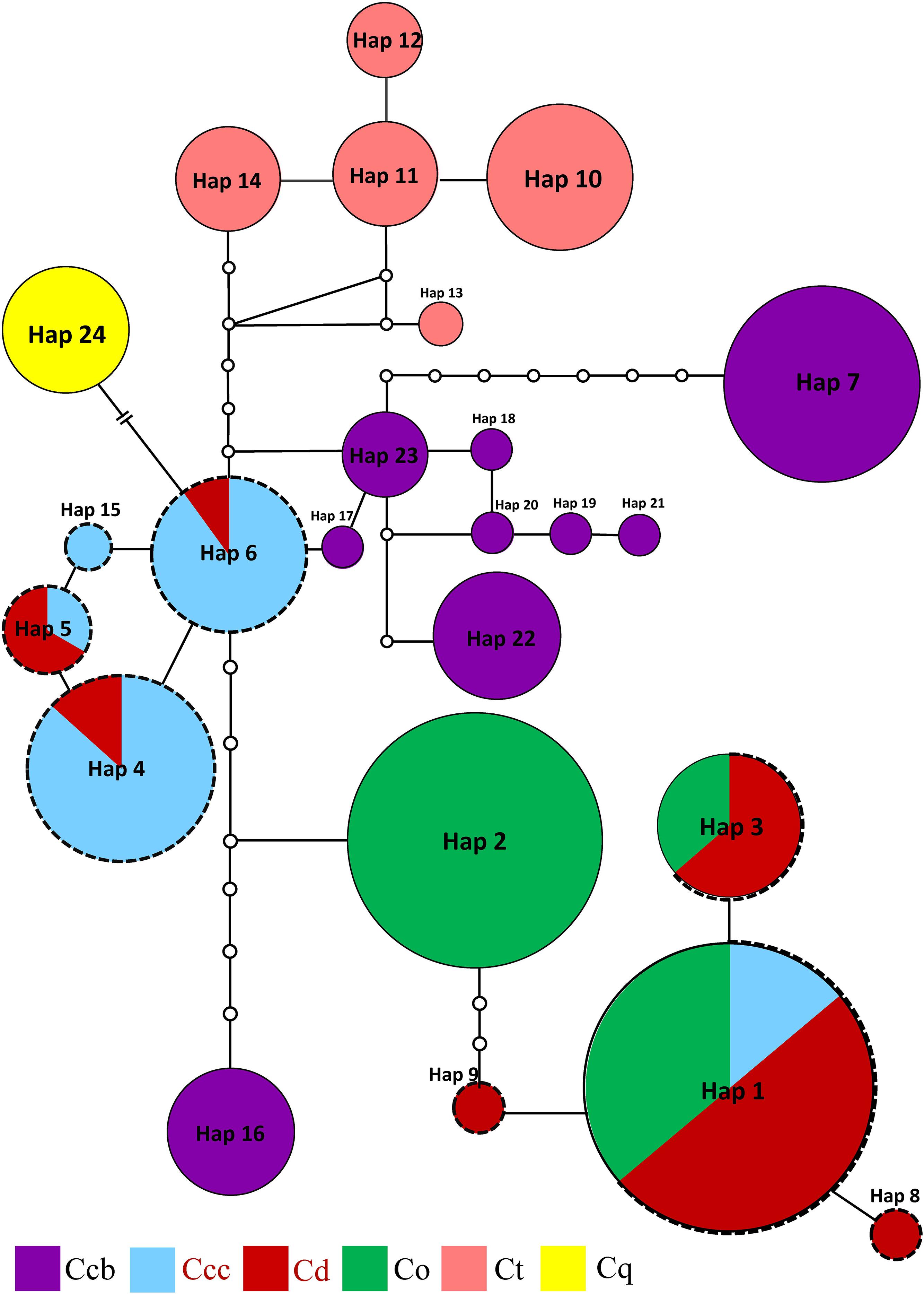
Figure 4. Parsimony network for all cpDNA haplotypes of C. chinensis var. brevisepala (Ccb), C. chinensis var. chinensis (Ccc), C. deltoidea (Cd), C. omeiensis (Co), C. teeta (Ct), and C. quinquefolia (Cq) species based on cpDNA data. The relative size of each circle corresponds to proportional to haplotype frequencies. The small hollow dots indicate hypothetical missing haplotypes. The network shown in different colors correspond to each species. The red letter and black dot line indicate that the sample is from cultivated.
Assuming substitution rates of 1.52 × 10–9 s/s/y, the divergence time of C. chinensis var. brevisepala populations and three species group (cultivated Huang-lian and C. omeiensis) was estimated as 7.6 Ma, with a 95% HPD confidence interval (95% CI) of 4.9–10.5 Ma (Figure 3). C. teeta diverged from the above four species earlier, about at 9.0 Ma (95% CI: 6.0−12.6 Ma). C. quinquefolia was firstly different from other species; the divergence time was estimated as 14.6 Ma (95% CI: 9.4−20.51 Ma). All results thus indicate that the divergence of Coptis species happened before Pleistocene (∼2.5–0.01 million years ago; Ma). Although no fossil record correction was used in this study, the results of the divergent time evaluation are similar to the previous study (Xiang et al., 2018).
Demographic History and Mismatch Analyses
A Bayesian skyline plot showed recent population size rise in the populations of cultivated Huang-lian (C. chinensis var. chinensis and C. deltoidea) and decline (C. chinensis var. brevisepala) or constant (C. omeiensis and C. teeta) in the populations of wild Coptis species (Figure 5). The cpDNA sequence mismatch analysis results for five species’ populations displayed a multimodal distribution pattern (Figure 6). There were nearly no significant SSD statistics and the HRag-value under the demographic expansion and spatial expansion models (Table 6). All these observations indicated the absence of an expansion event. However, whether the values of Tajima’s D and Fu and Li’s D∗ was positive or negative differed among species and populations (Table 2), which implied that demographic expansion existed in certain groups.
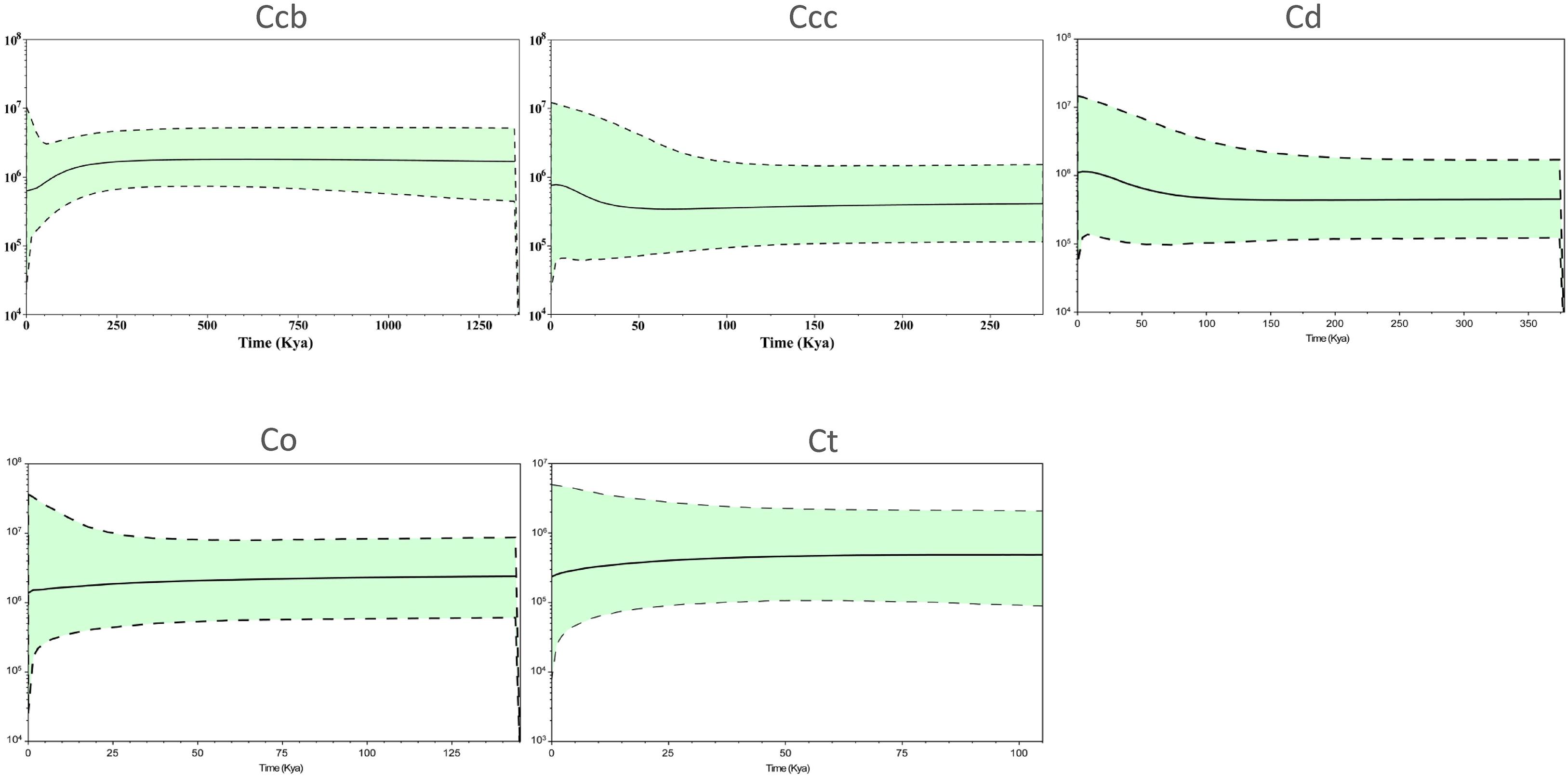
Figure 5. Bayesian skyline plot analysis for the effective population size over time for C. chinensis var. brevisepala (Ccb), C. chinensis var. chinensis (Ccc), C. deltoidea (Cd), C. omeiensis (Co), and C. teeta (Ct) species based on the cpDNA. The gray dash lines indicate 95 % confidence intervals.
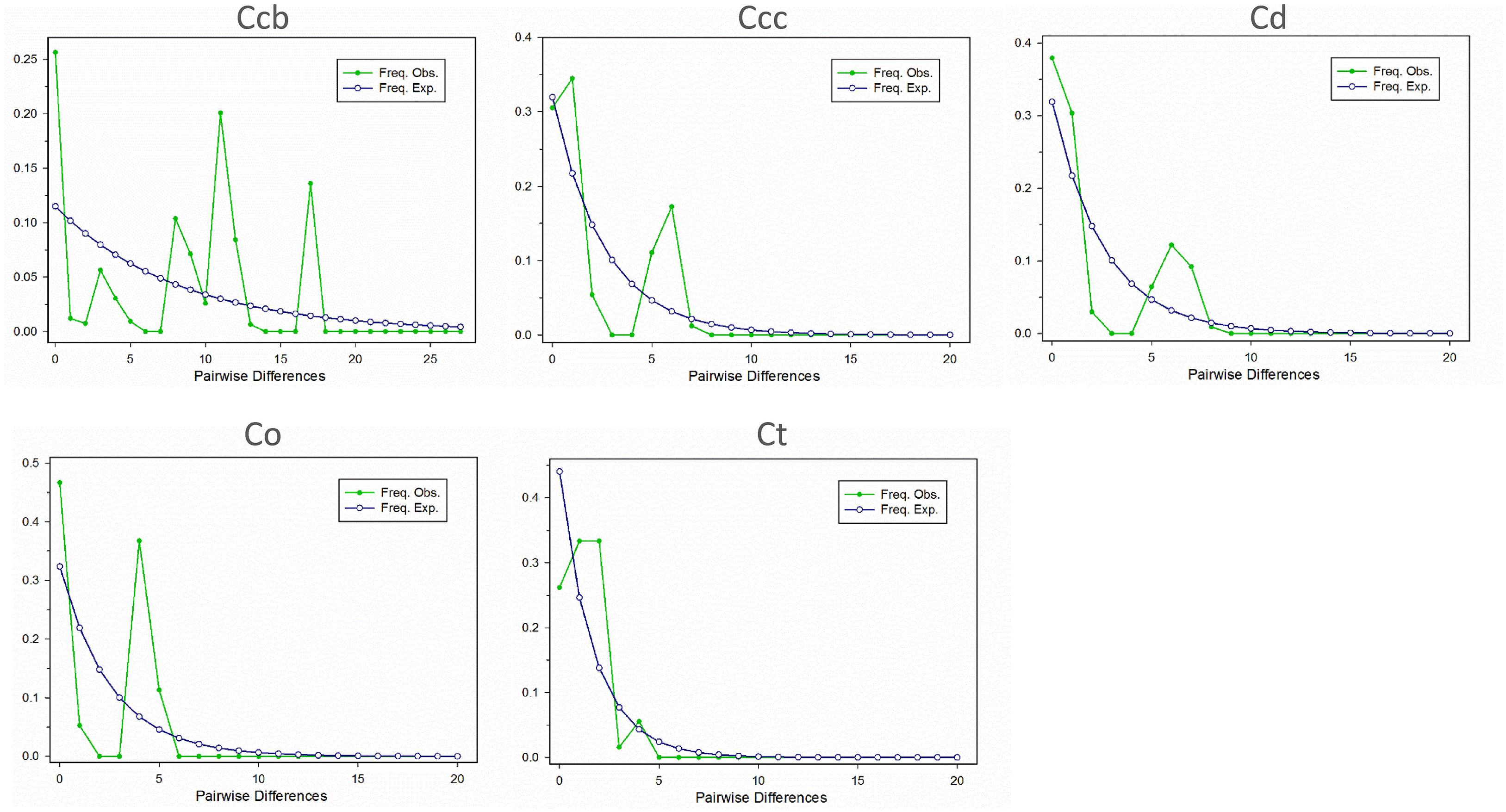
Figure 6. Mismatch distribution for cpDNA sequence data in C. chinensis var. brevisepala (Ccb), C. chinensis var. chinensis (Ccc), C. deltoidea (Cd), C. omeiensis (Co), and C. teeta (Ct) species, respectively. The line with hollow dot represents expected values and solid dot represents observed values.
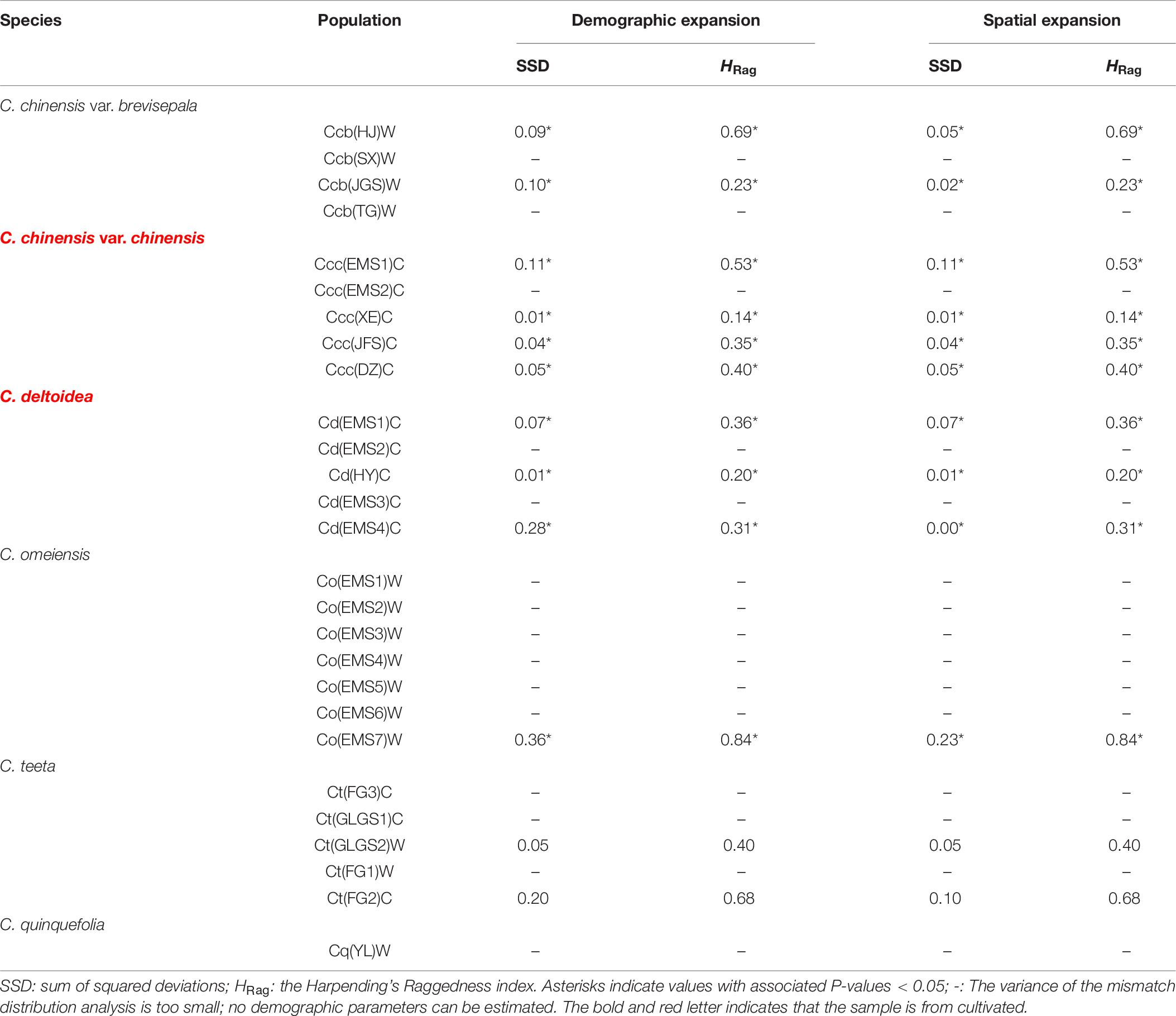
Table 6. Summary of mismatch distribution analysis (parameters of demographic and spatial expansion) of chloroplast DNA sequence data based on all populations of C. chinensis var. brevisepala, C. chinensis var. chinensis, C. deltoidea, C. omeiensis and C. teeta, respectively.
Historical Biogeography Inference
Ancestral ranges were obtained to infer vicariance and dispersal events in Coptis species by RASP analysis using plastid DNA data (Figure 7 and Table 7). Results support three dispersal events (node 44, 43 and 40) for the groups of two cultivated Huang-lian (C. chinensis var. chinensis and C. deltoidea) and C. omeiensis. The ancestor of the three species first colonized Sichuan (B), which was followed by one dispersal event, from Sichuan (B) to Chongqing and Hubei (F). Furthermore, dispersal + vicariance event was detected between the ancestor of the three species and C. chinensis var. brevisepala species on node 45, which indicates gene flow among them. However, distinctive vicariance signals (node 35 and 34) were observed in C. chinensis var. brevisepala, of which ancestor originated on Anhui (D), Jiangxi (G) and Guangxi (E), respectively, and several vicariance events occurring among these ancestor regions. Meanwhile, a vicariance event was also evident between C. teeta and the above four species (node 46), suggesting the independent origin of C. teeta.
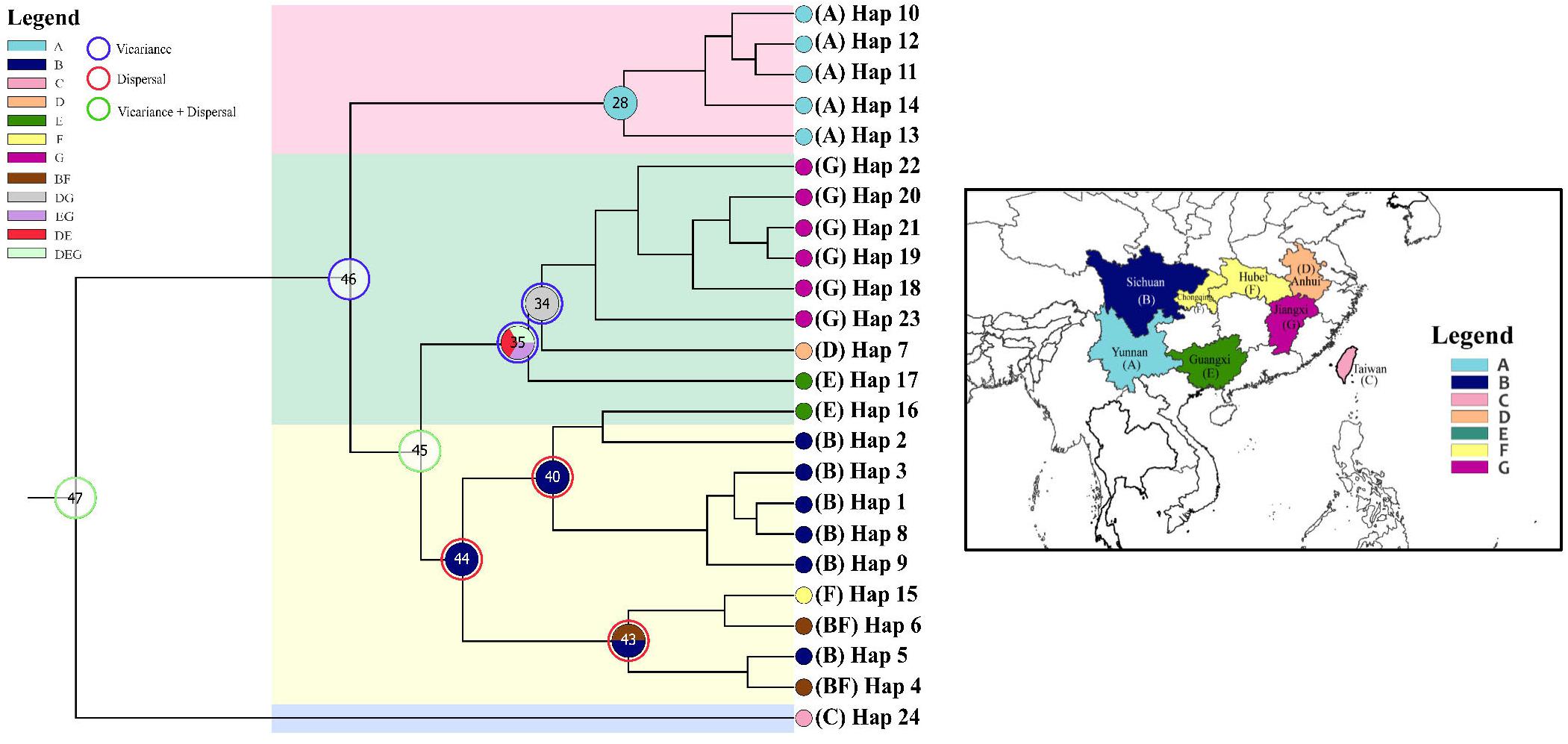
Figure 7. Ancestral distributions reconstructed and dispersal–vicariance analysis generated by RASP from the cpDNA data. Map showing seven geographical regions in colors as defined in RASP analysis. Pie charts on nodes display the relative probabilities of possible ancestral ranges.
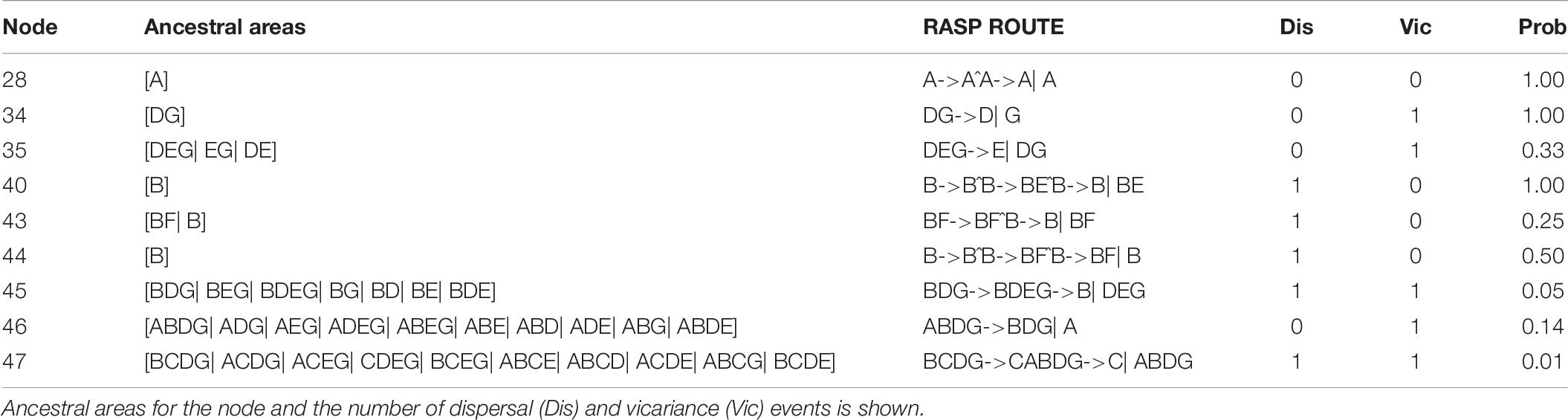
Table 7. The ancestral areas and dispersal-vicariance analysis inferred through RASP based on cpDNA data.
Discussion
The results of plant domestication were shaped by selection resulting from planting practices, human preferences, and agricultural environments, as well as the relevant population genetic processes following the reduction of the adequate population size (Smýkal et al., 2018). The domestication of wild plants always leads to genetic bottlenecks, and thus results in reduced genetic diversity due to founder effects and unintentional or intentional selections (Doebley et al., 2006). The analysis revealed that the genetic diversity of cultivated Huang-lian (C. chinensis var. chinensis and C. deltoidea) has not reduced compared with three wild Coptis species. The proportion of the population with unique haplotypes in cultivated Huang-lian was lower than that in wild Coptis species. However, the genetic diversity within populations of cultivated Huang-lian raised. The wild species and cultivated species have similar levels of genetic diversity. The loss of unique genetic composition in cultivated species are also to be found on medicinal plants of Scutellaria baicalensis (Yuan et al., 2010).
General agricultural large-scale planting practices significantly reduced the adequate population size of cultivated crops and allowed genetic drift (Smýkal et al., 2018). There is much evidence that domestication results in reduced genetic diversity in several cultivated crops and medicinal plants (Hollingsworth et al., 2005; Lam et al., 2010; Gao et al., 2017; Zhang et al., 2017). Although these bottlenecks affect the genetic diversity in the domestication process, the population sizes of the census will increase when the domesticated species are planted across extended areas (Smýkal et al., 2018). Furthermore, if the expanding population encounters a co-existing wild relative in a new area, the domesticated species and the wild species may have admixture or interbreeding, accompanied by genetic introgression events, and the domesticated population may have additional genetic components. This process might counteract the low genetic diversity caused by bottleneck effects and natural selection (Larson et al., 2014; Hazzouri et al., 2015; Mathew et al., 2015). Crops with significant hybridization or introgression, such as North African dates or citrus, and in outcrossing perennials crops, the cultivated populations may maintain high genetic diversity or increase relative to their wild relatives (Miller and Gross, 2011; Smýkal et al., 2018). A genetic bottleneck occurs during domestication, or during the founding events period, the composition of the gene pools may be further altered when the domesticated species moved away from the center of origin (Smýkal et al., 2018).
In China, C. chinensis var. chinensis is planted in large areas (0.32 million mus) (Wei and Zhang, 2006). C. deltoidea is only cultivated in Emeishan and adjacent regions, which cultivation areas have less than 5 thousand mus. Wild Coptis of C. omeiensis is mainly distributed on Emeishan in Sichuan province, overlapping with the distribution of C. deltoidea and C. chinensis var. chinensis. Therefore, a genetic material exchange may occur between Large-scale planting of cultivated Huang-lian and wild Coptis species, resulting in no significant decline in the genetic diversity of cultivated Huang-lian. In the cpDNA results of this study, cultivated Huang-lian and wild C. omeiensis shared multiple haplotypes, which indicates the existence of gene flows. The C. omeiensis has the lowest haplotype diversity. C. omeiensis has small population size and low natural fertility problems, which may cause low genetic diversity of plastid sequences (Huang, 2012).
Plant domestication affects not only the genetic variation of cultivated populations but also the structural patterns of genetic variation (Miller and Schaal, 2006). Assessing the genetic differentiation structure of cultivated Huang-lian and wild Coptis species, the genetic variation among intraspecies populations of cultivated Huang-lian showed lower population differentiation and a certain degree of homogenization. The wild Coptis species of C. quinquefolia and C. teeta have significant divergence from other species of Coptis. Wild Coptis species of C. chinensis var. brevisepala had the highest genetic variation among populations. These genetic distribution patterns indicate that the gene flow among populations of cultivated Huang-lian is higher relative to wild species of Coptis. Furthermore, C. chinensis var. chinensis is propagated by seeds and no stolon (Song et al., 2010) which might indicate widespread seed change among populations in different planting areas during extensive cultivation. Shi et al. (2008) used ISSR molecular markers to estimate genetic variation and gene flow among wild and cultivated populations of C. chinensis var. chinensis, showing a similar degree of genetic diversity and high gene flow between wild and cultivated populations (Shi et al., 2008). These results can support the present-day cultivated C. chinensis var. chinensis may have been introduced from a large number of wild ancestral seeds. Therefore, seeds from the primary gene pool include most of the genetic composition of the wild populations.
Moreover, the results of the demographic history analyses showed a recent population size rise in the populations of cultivated Huang-lian (Figure 5). This evidence also can infer that domesticated Huang-lian from abundant wild populations resources and have been domesticated and cultivated from multiple places through multiple events, resulting in a large initial population size of cultivated Huang-lian. Furthermore, historical biogeography inference also revealed multiple dispersal events in the groups of two cultivated Huang-lian (Figure 7 and Table 7). Additionally, mismatch distribution analysis showed a multimodal curve of pairwise differences which corresponds to no evident expansion and a recent bottleneck (Figure 6; Ray et al., 2003;Liao et al., 2010). However, neutrality tests were positive or negative differed among species and populations (Table 2). These analyses indicated that demographic expansion existed in specific groups and some groups experienced bottlenecks.
Compared with cultivated Huang-lian, wild relatives Coptis species have a high genetic differentiation among populations, indicating limited gene flow among populations. Wild Coptis species in China are facing habitat fragmentation crisis due to human over-exploitation and habitat demolition, causing some wild Coptis species are in an endangered crisis (Saha et al., 2015; Zhang et al., 2018). The Bayesian skyline plot also showed the recent population decline of certain wild Coptis species. However, the haplotype distribution results show that there are abundant unique haplotype resources in wild Coptis populations, which is very important for genetic improvement and conservation of domesticated species. Therefore, the intervention of conservation strategies is more important to maintain the genetic diversity of domesticated species.
The distribution of C. omeiensis overlaps with cultivated Huang-lian. At present, the cultivation practices of Huang-lian is mainly cultivated by traditional agriculture practices. Farmers plant Huang-lian in wild habitat, courtyard or semi-wild farming. Traditional agricultural practices systems can effectively maintain considerable genetic diversity resources has been proposed (Altieri, 2004; He et al., 2009; Guo et al., 2010; Yuan et al., 2010; Ren et al., 2018). These factors explain the close relationship between cultivated Huang-lian and C. omeiensis, and also indicate that C. deltoidea preserves considerable genetic resources of wild species. Moreover, it also explained the genetic diversity significant increase within the populations of cultivated Huang-lian. C. omeiensis was called as Emei wild Huang-lian and C. deltoidea was called as Emei domestic Huang-lian. Meanwhile, C. omeiensis was always used as an alternative Chinese herbal medicine for Huang-lian because their medicinal constituents are similar (Chen et al., 2017).
According to the ITS and cpDNA parsimony network information, C. chinensis var. chinensis is more closely related to C. chinensis var. brevisepala than C. omeiensis but does not form a monophyletic group. All haplotypes of cultivated Huang-lian and C. omeiensis were formed a monophyletic group (Figure 3), of which clustering the group contained all haplotypes of C. chinensis var. brevisepala except Hap16 of cpDNA haplotype. This similar pattern can also be obtained from the ITS parsimony network (Figure 2). Phylogenetic discernment between related taxa or between populations may be difficult because of the incomplete lineage sorting (Hollingsworth et al., 2011) or recently gene flow (Aguirre-Liguori et al., 2019). The sample contained in Hap16 corresponded to H10, H11, H140, H142, H148, H161 of ITS haplotypes (Figure 2 and Supplementary Table S2). The cultivated Huang-lian and C. chinensis var. brevisepala shared H30 of ITS haplotypes. From the ITS and cpDNA parsimony network showed that C. chinensis var. brevisepala has two genetic groups (Figures 2, 4). One genetic group is Hap16, corresponding to ITS H10, H11, H140, H142, H148, H161, and the second genetic group is the remaining Ccb haplotypes. The divergence time of C. chinensis var. brevisepala and C. chinensis var. chinensis (7.6 Ma) is earlier than the group of C. chinensis var. chinensis, C. deltoidea and C. omeiensis (Figure 3). According to biogeographic analysis, dispersal + vicariance event was detected between the ancestor of the cultivated Huang-lian and C. chinensis var. brevisepala species, which indicates gene flow among them and cause the two genetic group of C. chinensis var. brevisepala. However, several vicariance events occurring among C. chinensis var. brevisepala. C. chinensis var. brevisepala has abundant unique haplotypes, yet the restricted gene flow and vicariance among populations emphasize the importance of conserving genetic resources of wild relatives.
Understanding the natural genetic diversity and structure within and among populations of a species is essential for developing suitable conservation strategies to protect genetic resources and genetic improvements. Medicinal plants are a valuable source of herbal drug products. Proper assessment of medicinal plant genetic resources is useful information for developing conservation plans for protecting genetic diversity (Falk and Holsinger, 1991). Genetic information before domestication might have been retained in wild populations. According to haplotype distribution information, there are abundant unique genetic resources in wild relatives Coptis species. However, most wild relatives Coptis species of cultivated Huang-lian in China are facing an endangered crisis and are influenced by human activities. Thus, protection measures must be taken to avoid further reduction of wild Coptis species genetic resources. Currently, large-scale cultivation of cultivated Huang-lian becomes more wild populations go extinct. Restricted gene flow and population size decline in wild Coptis species highlight the importance of in situ and ex-situ conservation. Ex-situ conservation rerelease threatened species. Ex-situ focuses on the use of traditional and modern breeding techniques to maintain the genetic diversity of species. Regardless of the conservation strategies and resource management, the sustainable use of medicinal plant resources should be fully considered. The application of biotechnical approaches could help improve yield and modify the efficacy of medicinal plants (Kasagana and Karumuri, 2011; Chen et al., 2016). In this study, comprehensively evaluates the genetic resources of wild Coptis species and cultivated Huang-lian, insight into the genetic effects of long-term artificial selection on medicinal plants. This study provides practical perspectives to genetic resource conservation efforts and management for cultivated Huang-lian and relatives Coptis species.
Data Availability Statement
The datasets generated for this study can be found in the all information list on Supplementary Table S1.
Author Contributions
Q-JY, Y-CC, and L-QH conceived and designed the experiments. XW, X-QL, Y-ZK, X-LJ, J-HS, Z-YZ, Q-JY, and Y-CC performed the experiments. Y-ZK, X-LJ, Q-JY, and Y-CC analyzed the data. XW, X-QL, Y-ZK, X-LJ, J-HS, Z-YZ, Q-JY, and Y-CC contributed reagents, materials, and analysis tools. Y-ZK, Q-JY, Y-CC, and L-QH wrote the manuscript. XW, X-QL, Y-ZK, X-LJ, J-HS, Z-YZ, Q-JY, Y-CC, and L-QH conceived of the study, edited the manuscript, and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Key Research and Development Program of China (2017YFC1703700: 2017YFC1703704), the National Natural Science Foundation of China (NSFC: 81891010 and 81891014), Key Project at Central Government Level: The Ability Establishment of Sustainable Use for Valuable Chinese Medicine Resources (2060302) to Q-JY and L-QH, and by partial financing of NSYSU to Y-CC.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the editor OP and three reviewers for their constructive comments on the manuscript. The authors also thank Tsai-Wen Hsu and Shu-Jiau Chiou for help in Coptis quinquefolia collection in Taiwan.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00708/full#supplementary-material
References
Aguirre-Dugua, X., and González-Rodríguez, A. (2016). “Phylogeographical approaches to the study of plant domestication, with special emphasis on perennial plants,” in Ethnobotany of Mexico: Interactions of People and Plants in Mesoamerica, eds R. Lira and A. Casas (New York: Springer Press), 319–366. doi: 10.1007/978-1-4614-6669-7_13
Aguirre-Liguori, J. A., Gaut, B. S., Jaramillo-Correa, J. P., Tenaillon, M. I., Montes-Hernández, S., García-Oliva, F., et al. (2019). Divergence with gene flow is driven by local adaptation to temperature and soil phosphorus concentration in teosinte subspecies (Zea mays parviglumis and Zea mays mexicana). Mol. Ecol. 28, 2814–2830.
Akaike, H. (1974). A new look at the statistical model identification. IEEE Trans. Auto Cont. 19, 716–723. doi: 10.1109/TAC.1974.1100705
Allaby, R. G., Ware, R. L., and Kistler, L. (2019). A re−evaluation of the domestication bottleneck from archaeogenomic evidence. Evol. Appl. 12, 29–37. doi: 10.1111/eva.12680
Altieri, M. A. (2004). Linking ecologists and traditional farmers in the search for sustainable agriculture. Front. Ecol. Evol 2:2. doi: 10.1890/1540-92952004002[0035:LEATFI]2.0.CO;2
Avise, J. C. (2000). Phylogeography: The History and Formation of Species. Cambridge, MA: Harvard University Press.
Bodeker, G., Bhat, K. K. S., Burley, J., and Vantimme, P. (1997). Medicinal Plants for Forest Conservation and Health Care. Rome: Food & Agriculture Organization of the United Nations.
Caicedo, A. L., and Schaal, B. A. (2004). Population structure and phylogeography of Solanum pimpinellifolium inferred from a nuclear gene. Mol. Ecol. 13, 1871–1882. doi: 10.1111/j.1365-294X.2004.02191.x
Chen, D. X., Li, L. Y., Peng, R., and Qu, X. Y. (2006). Genetic diversity of Coptis chinensis germplasm based on ISSR analysis. China J. Chin. Mater. Med. 31, 1937–1940.
Chen, H., Fan, G., and He, Y. (2017). Species evolution and quality evaluation of four Coptis herbal medicinal materials in Southwest China. 3 Biotech 7:62. doi: 10.1007/s13205-017-0679-8
Chen, S. L., Yu, H., Luo, H. M., Wu, Q., Li, C. F., and Steinmetz, A. (2016). Conservation and sustainable use of medicinal plants: problems, progress, and prospects. Chin. Med. 11:37. doi: 10.1186/s13020-016-0108-7
Chen, Y. Y., Bao, Z. X., Qu, Y., and Li, Z. Z. (2014). Genetic variation in cultivated populations of Gastrodia elata, a medicinal plant from central China analyzed by microsatellites. Genet. Resour. Crop. Evol. 61, 1523–1532. doi: 10.1007/s10722-014-0127-0
China, P. (2015). Pharmacopoeia of the People’s Republic of China. Beijing: China Medical Science and Technology Press.
Clement, M., and Crandall, K. P. D. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Doebley, J. F., Gaut, B. S., and Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. doi: 10.1016/j.cell.2006.12.006
Doyle, J., and Doyle, J. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Drummond, A. J., and Rambaut, A. (2007). BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50.
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Falk, K. E., and Holsinger, D. F. (1991). Genetics and Conservation of Rare Plants. New York, NY: Oxford University Press.
Farris, J. S., Källersjö, M., Kluge, A. G., and Bult, C. (1994). Testing significance of incongruence. Cladistics 10, 315–319. doi: 10.1111/j.1096-0031.1994.tb00181.x
Felsenstein, J. (1993). PHYLIP (Phylogeny Inference Package), Version 3.5c. Philadelphia, PL: Joseph Felsenstein.
Fu, D., and Orbélia, R. R. (2001). “Coptis,” in Flora of China, Vol. 6, eds Z. Y. Wu and P. H. Raven (Beijing: Science Press), 305–307.
Fu, Y. X., and Li, W. H. (1993). Statistical tests of neutrality of mutations. Genetics 133, 693–709. doi: 10.0000/PMID8454210
Gao, Y., Yin, S., Wu, L., Dai, D., Wang, H., Liu, C., et al. (2017). Genetic diversity and structure of wild and cultivated Amorphophallus paeoniifolius populations in southwestern China as revealed by RAD-seq. Sci. Rep. 7:14183. doi: 10.1038/s41598-017-14738-6
Group, C. P. W. (2009). A DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A. 106, 12794–12797. doi: 10.1073/pnas.0905845106
Guo, H. B., Cui, X. M., An, N., and Cai, G. P. (2010). Sanchi ginseng (Panax notoginseng (Burkill) F. H. Chen) in China: distribution, cultivation and variations. Genet. Resour. Crop Evol. 57, 453–460. doi: 10.1007/s10722-010-9531-2
Harpending, H. (1994). Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum. Biol. 66, 591–600. doi: 10.1038/hdy.1994.122
Haudry, A., Cenci, A., Ravel, C., Bataillon, T., Brunel, D., Poncet, C., et al. (2007). Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 24, 1506–1517. doi: 10.1093/molbev/msm077
Hazzouri, K. M., Flowers, J. M., Visser, H. J., Khierallah, H. S. M., Rosas, U., Pham, G. M., et al. (2015). Whole genome re-sequencing of date palms yields insights into diversification of a fruit tree crop. Nat. Commun. 6:8824. doi: 10.1038/ncomms9824
He, J., Chen, L., Si, Y., Huang, B., Ban, X., and Wang, Y. (2009). Population structure and genetic diversity distribution in wild and cultivated populations of the traditional Chinese medicinal plant Magnolia officinalis subsp. biloba (Magnoliaceae). Genetica 135, 233–243. doi: 10.1007/s10709-008-9272-8
He, Y., Hou, P., Fan, G., Arain, S., and Peng, C. (2014). Comprehensive analyses of molecular phylogeny and main alkaloids for Coptis (Ranunculaceae) species identification. Biochem. Syst. Ecol. 56, 88–94. doi: 10.1016/j.bse.2014.05.002
Hollingsworth, P. M., Dawson, I. K., Goodall-Copestake, W. P., Richardson, J. E., Weber, J. C., Montes, C. S., et al. (2005). Do farmers reduce genetic diversity when they domesticate tropical trees? A case study from Amazonia. Mol. Ecol. 14, 497–501. doi: 10.1111/j.1365-294X.2005.02431.x
Hollingsworth, P. M., Graham, S. W., and Little, D. P. (2011). Choosing and using a plant DNA barcode. PLoS One 6:e19254. doi: 10.1371/journal.pone.0019254
Huang, H. (2011). Plant diversity and conservation in China: planning a strategic bioresource for a sustainable future. Bot. J. Linn. Soc. 166, 282–300. doi: 10.1111/j.1095-8339.2011.01157.x
Huang, J., Chen, Z., Xia, Z., and Shang, H. (2013). Chromosome karyotypes of Coptis species in China. Acta Bot. Bor Occid. Sinica 33, 931–938.
Huang, X., Kurata, N., Wang, Z. X., Wang, A., Zhao, Q., Zhao, Y., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Hyten, D. L., Qijian, S., Youlin, Z., Ik-Young, C., Nelson, R. L., Costa, J. M., et al. (2006). Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. U.S.A. 103, 16666–16671. doi: 10.1073/pnas.0604379103
Innis, M. A., Gelfand, D. H., Sninsky, J. J., and White, T. J. (1990). PCR Protocols: A Guide to Methods and Applications. London: Academic Press.
Kasagana, V. N., and Karumuri, S. S. (2011). Conservation of medicinal plants (past, present & future trends). J. Pharm. Sci. Res. 3:1378.
Kovach, M. J., and Mccouch, S. R. (2008). Leveraging natural diversity: back through the bottleneck. Curr. Opin. Plant Biol. 11, 193–200. doi: 10.1016/j.pbi.2007.12.006
Kuchta, S. R., and Meyer, D. (2001). A genealogical view of geographical variation. Mol. Ecol. 10, 2569–2576. doi: 10.1046/j.0962-1083.2001.01396.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lam, H. M., Xu, X., Liu, X., Chen, W., Yang, G., Wong, F.-L., et al. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42, 1053–1059. doi: 10.1038/ng.715
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Larson, G., Piperno, D. R., Allaby, R. G., Purugganan, M. D., Andersson, L., Arroyo-Kalin, M., et al. (2014). Current perspectives and the future of domestication studies. Proc. Natl. Acad. Sci. U.S.A. 111, 6139–6146. doi: 10.1073/pnas.1323964111
Li, D. Z., Gao, L. M., Li, H. T., Wang, H., Ge, X. J., Liu, J.-Q., et al. (2011). Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. U.S.A. 108, 19641–19646. doi: 10.1073/pnas.1104551108
Li, X., Chen, Y., Lai, Y., Yang, Q., Hu, H., and Wang, Y. (2015). Sustainable utilization of traditional chinese medicine resources: systematic evaluation on different production modes. Evid. Based Comp. Alternat. Med. 2015:218901. doi: 10.1155/2015/218901
Li, X., Song, L., Dai, C., Tang, H., Wang, H., and Yan, Z. (2010). Breeding system, morphological and anatomical characteristics of Coptis deltoidea. China J. Chin. Mat. Med. 35, 1789–1794. doi: 10.4268/cjcmm20101402
Liao, P. C., Kuo, D. C., Lin, C. C., Ho, K. C., Lin, T. P., and Hwang, S.-Y. (2010). Historical spatial range expansion and a very recent bottleneck of Cinnamomum kanehirae Hay. (Lauraceae) in Taiwan inferred from nuclear genes. BMC Evol. Biol. 10:124. doi: 10.1186/1471-2148-10-124
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Loera-Sánchez, M., Studer, B., and Kölliker, R. (2019). DNA-based assessment of genetic diversity in grassland plant species: challenges, approaches, and applications. Agron 9:881. doi: 10.3390/agronomy9120881
Londo, J. P., Chiang, Y. C., Hung, K. H., Chiang, T. Y., and Schaal, B. A. (2006). Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. U.S.A. 103:9578. doi: 10.1073/pnas.0603152103
Lv, X., Li, Y., Tang, C., Zhang, Y., Zhang, J., and Fan, G. (2016). Integration of HPLC-based fingerprint and quantitative analyses for differentiating botanical species and geographical growing origins of Rhizoma coptidis. Pharm. Biol. 54, 3264–3271. doi: 10.1080/13880209.2016.1223699
Mathew, L. S., Seidel, M. A., George, B., Mathew, S., Spannagl, M., Haberer, G., et al. (2015). A Genome-wide survey of date palm cultivars supports two major subpopulations in Phoenix dactylifera. G3 (Bethesda) 5, 1429–1438. doi: 10.1534/g3.115.018341
Miller, A. J., and Gross, B. L. (2011). From forest to field: perennial fruit crop domestication. Am. J. Bot. 98, 1389–1414. doi: 10.3732/ajb.1000522
Miller, A. J., and Schaal, B. A. (2006). Domestication and the distribution of genetic variation in wild and cultivated populations of the Mesoamerican fruit tree Spondias purpurea L. (Anacardiaceae). Mol. Ecol. 15, 1467–1480. doi: 10.1111/j.1365-294X.2006.02834.x
Mukherjee, D., and Chakraborty, S. (2019). Coptis Teeta: conservation and cultivation practice-A rare medicinal plant on Earth. Curr. Invest. Agric. Curr. Res. 6, 845–851. doi: 10.32474/CIACR.2019.06.000244
Niazi, I. A. K., Rauf, S., da Silva, J. A. T., and Munir, H. (2015). Comparison of teosinte (Zea mexicana L.) and inter-subspecific hybrids (Zea mays L.× Zea mexicana) for high forage yield under two sowing regimes. Crop Pasture Sci. 66, 49–61. doi: 10.1071/CP14155
Olsen, K. M., and Gross, B. L. (2008). Detecting multiple origins of domesticated crops. Proc. Natl. Acad. Sci. U.S.A. 105, 13701–13702. doi: 10.1073/pnas.0807439105
Pandit, M. K., and Babu, C. R. (1993). Cytology and taxonomy of Coptis teeta Wall. (Ranunculaceae). Bot. J. Linn. Soc. 111, 371–378. doi: 10.1006/bojl.1993.1026
Payum, T. (2017). Distribution, ethnobotany, pharmacognosy and phytoconstituents of Coptis teeta Wall.: a highly valued and threatened medicinal plant of Eastern Himalayas. Pharmacogn. J. 9, s28–s34. doi: 10.5530/pj.2017.6s.154
Posada, D. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Rambaut, A. (2012). FigTree v. 1. 4. Molecular Evolution, Phylogenetics and Epidemiology. Edinburgh: University of Edinburgh.
Rambaut, A., and Drummond, A. (2009). Tracer 1.5. 0. MCMC Trace Analysis Tool. Available online at: http://tree.bio.ed.ac.uk/software/tracer/ (accessed February 19, 2018)Google Scholar
Ray, N., Currat, M., and Excoffier, L. (2003). Intra-deme molecular diversity in spatially expanding populations. Mol Biol Evol 20, 76–86. doi: 10.1093/molbev/msg009
Ren, W., Hu, L., Guo, L., Zhang, J., Tang, L., Zhang, E., et al. (2018). Preservation of the genetic diversity of a local common carp in the agricultural heritage rice-fish system. Proc Natl Acad Sci U S A 115, E546–E554. doi: 10.1073/pnas.1709582115
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Saha, D., Ved, D., Haridasan, K., and Ravikumar, K. (2015). Coptis Teeta. The IUCN Red List of Threatened Species 2015 e.T50126583A50131320.
Sang, T., Crawford, D. J., and Stuessy, T. F. (1997). Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 84, 1120–1136. doi: 10.2307/2446155
Schippmann, U., Leaman, D. J., and Cunningham, A. (2002). “Impact of cultivation and gathering of medicinal plants on biodiversity: global trends and issues (Case Study No. 7),” in Biodiversity and the Ecosystem Approach in Agriculture, Forestry and Fisheries, Satellite Event Session on the Occasion of the 9th Regular Session of the Commission on “Genetic Resources for Food and Agriculture”, ed. FAO (Rome: FAO Document Repository of United Nations).
Selvam, A. B. D., and India, B. S. O. (2012). Pharmacognosy of Negative Listed Plants. Kolkata: Botanical Survey of India.
Shi, W., Yang, C. F., Chen, J. M., and Guo, Y. H. (2008). Genetic variation among wild and cultivated populations of the Chinese medicinal plant Coptis chinensis (Ranunculaceae). Plant Biol. 10, 485–491. doi: 10.1111/j.1438-8677.2008.00035.x
Smýkal, P., Nelson, M., Berger, J., and Von Wettberg, E. (2018). The impact of genetic changes during crop domestication on healthy food development. Agronomy 8:26. doi: 10.3390/agronomy8030026
Song, L., Wang, H., He, H., Dai, C., Li, X., and Dong, G. (2010). Life cycle and characteristics of reproduction and ecology of the endangered plant Coptis omeiensis. Chin. Bull. Bot. 45, 444–450. doi: 10.3969/j.issn.1674-3466.2010.04.007
Swofford, D. (2002). PAUP∗ Phylogeny Analysis Using Parsimony (∗ and other methods), version 40b10. Sunderland, MA: Sinauer Association, Inc.
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. doi: 10.0000/PMID2513255
Tamura, M. (1995). “Coptis,” in Die Natürlichen Pflanzenfamilien, 2nd Edn, ed. P. Hiepko (Berlin: Duncker and Humblot), 67.
Tan, H. L., Chan, K. G., Pusparajah, P., Duangjai, A., Saokaew, S., Khan, T. M., et al. (2016). Rhizoma Coptidis: A potential cardiovascular protective agent. Front. Pharmacol. 7:362. doi: 10.3389/fphar.2016.00362
Tenaillon, M. I., U’ren, J., Tenaillon, O., and Gaut, B. S. (2004). Selection versus demography: a multilocus investigation of the domestication process in maize. Mol. Biol. Evol. 21, 1214–1225. doi: 10.1093/molbev/msh102
Wang, D. Q., and Peng, H. S. (2008). The vicissitude of the historically famous medicinal herb Xuan Huanglian (Coptis chinensis var. brevisepala). Chin. J. Med. Hist. 38, 137–139.
Wang, J., Wang, L., Lou, G. H., Zeng, H. R., Hu, J., Huang, Q. W., et al. (2019). Coptidis Rhizoma: a comprehensive review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. Pharm. Biol. 57, 193–225. doi: 10.1080/13880209.2019.1577466
Wei, J., and Zhang, L. (2006). Chinese Herbal Drug Industry: Past, Present and Future. 6000 Broken Sound Parkway NW, Suite 300. Boca Raton, FL: Taylor & Francis Group, LLC.
Wolfe, K. H., Li, W. H., and Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. U.S.A. 84, 9054–9058. doi: 10.1073/pnas.84.24.9054
Xia, X., and Xie, Z. (2001). DAMBE: Software package for data analysis in molecular biology and evolution. J. Hered. 30, 1720–1728. doi: 10.1093/molbev/mst064
Xiang, G., Fu, T., and Fan, Q. (2010). Chromosome characteristics of three Coptis species. China J. Chin. Mat. Med. 35, 1367–1371. doi: 10.4268/cjcmm20101101
Xiang, K. L., Erst, A. S., Xiang, X. G., Jabbour, F., and Wang, W. (2018). Biogeography of Coptis salisb. (Ranunculales, Ranunculaceae, Coptidoideae), an Eastern Asian and North American genus. BMC Evol. Biol. 18:74. doi: 10.1186/s12862-018-1195-0
Xiang, K. L., Wu, S. D., Yu, S. X., Liu, Y., Jabbour, F., Erst, A. S., et al. (2016b). The First Comprehensive phylogeny of Coptis (Ranunculaceae) and its implications for character evolution and classification. PLoS One 11:e0153127. doi: 10.1371/journal.pone.0153127
Xiao, P. G. (1991). “The Chinese approach to medicinal plants. Their utilization and conservation,” in Conservation of Medicinal Plants, eds O. Akerele, V. Heywood, and H. Synge (Cambridge: University Press), 305–313.
Xiong, F. Y., Ma, Y. T., Yan, Z. Y., Xu, R. C., Ma, Q., Wu, Q. H., et al. (2011). The present research situation of conservation biology on plants of coptis in China. Pharm. Clin. Chin. Mater. Med. 2, 11–14.
Yamasaki, M., Wright, S. I., and Mcmullen, M. D. (2007). Genomic screening for artificial selection during domestication and improvement in maize. Ann. Bot. 100, 967–973. doi: 10.1093/aob/mcm173
Yang, S. (1998). The Divine Farmer’s Materia Medica: A Translation of the Shen Nong Ben Cao Jing. Boulder, CO: Blue Poppy Press.
Yu, Y., Harris, A. J., Blair, C., and He, X. (2015). RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol. 87, 46–49. doi: 10.1016/j.ympev.2015.03.008
Yu, Y. Y., Wang, B. C., Peng, L., Wang, J. B., and Zeng, C. (2006). Advances in pharmacological studies of Coptis chinensis. J. Chongqing 29, 107–111.
Yuan, Q. J., Zhang, Z. Y., Hu, J., Guo, L. P., Shao, A. J., and Huang, L. Q. (2010). Impacts of recent cultivation on genetic diversity pattern of a medicinal plant, Scutellaria baicalensis (Lamiaceae). BMC Genet. 11:29. doi: 10.1186/1471-2156-11-29
Zhang, J., Wang, X., Yao, J., Li, Q., Liu, F., Yotsukura, N., et al. (2017). Effect of domestication on the genetic diversity and structure of Saccharina japonica populations in China. Sci. Rep. 7:42158. doi: 10.1038/srep42158
Zhang, L., and Zhang, X. (2005). Population characteristics of Coptis chinensis var. brevisepala in Anhui Province and its endangering mechanism. Chin. J. Appl. Ecol. 16, 1394–1398. doi: 10.1360/aps040037
Zhang, Y., Dong, J., Feng, Y., Wang, Z., and Li, P. (2018). The first complete chloroplast genome of Coptis quinquesecta, a critically endangered medicinal plant in China. Mitochondrial DNA Part B 3, 370–372. doi: 10.1080/23802359.2018.1450684
Keywords: cultivated Huang-lian, wild Coptis species, plastid DNA, genetic diversity, phylogeography
Citation: Wang X, Liu X-Q, Ko Y-Z, Jin X-L, Sun J-H, Zhao Z-Y, Yuan Q-J, Chiang Y-C and Huang L-Q (2020) Genetic Diversity and Phylogeography of the Important Medical Herb, Cultivated Huang-Lian Populations, and the Wild Relatives Coptis Species in China. Front. Genet. 11:708. doi: 10.3389/fgene.2020.00708
Received: 05 December 2019; Accepted: 10 June 2020;
Published: 03 July 2020.
Edited by:
Octavio Salgueiro Paulo, University of Lisbon, PortugalReviewed by:
Isabel Marques, University of Lisbon, PortugalAllison Miller, Saint Louis University, United States
Shilong Chen, Northwest Institute of Plateau Biology (CAS), China
Copyright © 2020 Wang, Liu, Ko, Jin, Sun, Zhao, Yuan, Chiang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing-Jun Yuan, yuanqingjun@icmm.ac.cn; Yu-Chung Chiang, yuchung@mail.nsysu.edu.tw; Lu-Qi Huang, huangluqi01@126.com
†These authors have contributed equally to this work
 Xin Wang
Xin Wang Xiao-Quang Liu
Xiao-Quang Liu Ya-Zhu Ko
Ya-Zhu Ko Xiao-Lei Jin
Xiao-Lei Jin Jia-Hui Sun
Jia-Hui Sun Zhen-Yu Zhao
Zhen-Yu Zhao Qing-Jun Yuan
Qing-Jun Yuan Yu-Chung Chiang
Yu-Chung Chiang Lu-Qi Huang
Lu-Qi Huang