The role of the plasmid-mediated fluoroquinolone resistance genes as resistance mechanisms in pediatric infections due to Enterobacterales
- 1Pediatrics, Emory University School of Medicine, Atlanta, GA, United States
- 2Department of Pediatrics, Children’s Healthcare of Atlanta, Atlanta, GA, United States
- 3Pediatrics, Rush University Medical Center, Chicago, IL, United States
- 4Research Service, Louis Stokes Cleveland Department of Veterans Affairs Medical Center, Cleveland, OH, United States
- 5Molecular Biology, and Microbiology, Case Western Reserve University School of Medicine, Cleveland, OH, United States
- 6Medicine, Rush University Medical Center, Chicago, IL, United States
- 7Biostatistics, University of Illinois at Chicago, Chicago, IL, United States
- 8Pediatrics, Loyola University Medical Center, Maywood, IL, United States
- 9Medicine, Case Western Reserve University School of Medicine, Cleveland, OH, United States
- 10Microbiology, Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, IL, United States
- 11Pathology, Northwestern Feinberg School of Medicine, Chicago, IL, United States
- 12Department of Medicine, Cook County Health, Chicago, IL, United States
- 13Biochemistry, Pharmacology, Proteomics and Bioinformatics, Case Western Reserve University School of Medicine, Cleveland, OH, United States
- 14Case Western Reserve University (CWRU)-Cleveland VA Medical Center (VAMC) Center for Antimicrobial Resistance and Epidemiology (Case VA CARES), Cleveland, OH, United States
Introduction: Fluoroquinolones (FQs) are not commonly prescribed in children, yet the increasing incidence of multidrug-resistant (MDR) Enterobacterales (Ent) infections in this population often reveals FQ resistance. We sought to define the role of FQ resistance in the epidemiology of MDR Ent in children, with an overall goal to devise treatment and prevention strategies.
Methods: A case–control study of children (0–18 years) at three Chicago hospitals was performed. Cases had infections by FQ-susceptible, β-lactamase-producing (bla) Ent harboring a non- or low-level expression of PMFQR genes (PMFQS Ent). Controls had FQR infections due to bla Ent with expressed PMFQR genes (PMFQR Ent). We sought bla genes by PCR or DNA (BD Max Check-Points assay®) and PMFQR genes by PCR. We performed rep-PCR, MLST, and E. coli phylogenetic grouping. Whole genome sequencing was additionally performed on PMFQS Ent positive isolates. Demographics, comorbidities, and device, antibiotic, and healthcare exposures were evaluated. Predictors of infection were assessed.
Results: Of 170 β-lactamase-producing Ent isolates, 85 (50%) were FQS; 23 (27%) had PMFQR genes (PMFQS cases). Eighty-five (50%) were FQR; 53 (62%) had PMFQR genes (PMFQR controls). The median age for children with PMFQS Ent and PMFQR Ent was 4.3 and 6.2 years, respectively (p = NS). Of 23 PMFQS Ent, 56% were Klebsiella spp., and of 53 PMFQR Ent, 76% were E. coli. The most common bla and PMFQR genes detected in PMFQS Ent were blaSHV ESBL (44%) and oqxAB (57%), and the corresponding genes detected in PMFQR Ent were blaCTX-M-1-group ESBL (79%) and aac(6’)-Ib-cr (83%). Whole genome sequencing of PMFQS Ent revealed the additional presence of mcr-9, a transferable polymyxin resistance gene, in 47% of isolates, along with multiple plasmids and mobile genetic elements propagating drug resistance. Multivariable regression analysis showed that children with PMFQS Ent infections were more likely to have hospital onset infection (OR 5.7, 95% CI 1.6–22) and isolates containing multiple bla genes (OR 3.8, 95% CI 1.1–14.5). The presence of invasive devices mediated the effects of healthcare setting in the final model. Differences in demographics, comorbidities, or antibiotic use were not found.
Conclusions: Paradoxically, PMFQS Ent infections were often hospital onset and PMFQR Ent infections were community onset. PMFQS Ent commonly co-harbored multiple bla and PMFQR genes, and additional silent, yet transferrable antibiotic resistance genes such as mcr-9, affecting therapeutic options and suggesting the need to address infection prevention strategies to control spread. Control of PMFQS Ent infections will require validating community and healthcare-based sources and risk factors associated with acquisition.
Introduction
Multidrug-resistant (MDR) Enterobacterales (Ent) infections are associated with significant morbidity and mortality and are a growing concern among children and healthy populations in the US (Logan et al., 2014; Logan et al., 2015b). This dramatic rise in resistance is globally attributed mainly to extended-spectrum β-lactamase (ESBL)-producing Enterobacterales and carbapenem-resistant Enterobacterales (CRE) (Logan et al., 2015a; Lukac et al., 2015; Logan and Weinstein, 2017). However, increasing resistance to several other classes of antibiotics is co-occurring, including to the fluoroquinolones (FQs), a class of antibiotics that are not commonly prescribed in children, yet the increasing incidence of MDR Ent infections in the pediatric population often reveals FQ resistance (Medernach and Logan, 2018).
Among MDR Enterobacterales, Escherichia coli harboring blaCTX-M are the most common ESBL-producing Enterobacterales circulating, and the bacteria often possess resistance genes to other important antibiotic classes including fluoroquinolones, aminoglycosides, trimethoprim-sulfamethoxazole, and tetracyclines (Cantón and Coque, 2006; Price et al., 2013). In adults, fluoroquinolone resistance (FQR) in Gram-negative bacteria has been linked to both chromosomal and plasmid-mediated resistance mechanisms and most likely associated with the increase in use of these antibiotics during the 1980s (Robicsek et al., 2006; Mathers et al., 2015). However, there has been an increasing prevalence of global fluoroquinolone-resistant clones of E. coli, including among healthy populations, despite demonstrated decreases in fluoroquinolone use (Tchesnokova et al., 2018; Pitout et al., 2022).
Phenotypic expression of FQR among Enterobacterales often involves chromosomal mechanisms of resistance, specifically mutations in gyrA and parC genes of the quinolone resistance determining region (QRDR) in E. coli, Klebsiella spp., and Enterobacter spp (Chen et al., 2012). Plasmid-mediated mechanisms of FQR often have lower levels of expression, and therefore the presence of these transmissible genetic mechanisms of resistance among Enterobacterales isolates may be underrecognized when chromosomal resistance mechanisms are not present (Diancourt et al., 2005; Chen et al., 2012).
As fluoroquinolones are uncommonly prescribed in children, the reasons for the increases in FQR Enterobacterales among children are unclear. Studies assessing FQR in Enterobacterales among children are limited, including the analysis of plasmid-mediated (PM) resistance mechanisms associated with the FQR phenotype (PMFQR) and the FQS phenotype (PMFQS) (Medernach and Logan, 2018; Logan et al., 2019b). Therefore, we examined this population to determine which children had a higher likelihood of infections with Enterobacterales resistant to β-lactam antibiotics, yet variable susceptibilities to fluoroquinolone antibiotics, and determined the genotypes, host factors, and exposures associated with infection. We seek to define the role of FQR in the epidemiology of MDR Enterobacterales colonization and infection in children, with an overall goal to devise effective treatment and prevention strategies.
Methods
Study setting
Isolates were obtained from three academic centers in the metropolitan Chicago area. Hospital A is a 115-bed children’s hospital within a tertiary care academic medical center that has a step-down care unit, a pediatric ward, a mother–newborn infant unit, and cardiac, neonatal, and pediatric intensive care units (CICU, NICU, and PICU). Hospital B is a 125-bed children’s hospital within an academic medical center, and contains general pediatric and newborn infant wards, as well as a NICU and PICU. Hospital C is a freestanding children’s academic medical center with 288 beds that provides complex quaternary services, such as pediatric solid organ and bone marrow transplantation.
Descriptive study design
Study population
This study included patients aged 0 to 18.99 years who had clinical cultures positive for Enterobacterales with extended-spectrum cephalosporin and/or carbapenem resistance and the suspected presence of a β-lactamase gene based on clinical microbiology laboratory testing results. Isolates were grouped and further characterized based on susceptibility to fluoroquinolones. Infections were diagnosed between 1 January 2011 and 31 December 2016, and only the first infection episode per pediatric patient was included. The study was approved by the institutional review boards of the three participating institutions and the need for informed consent was waived by all institutions.
Antibiotic susceptibility testing in Enterobacterales
The clinical microbiology laboratories of Hospitals A–C phenotypically analyzed presumed β-lactamase-producing Enterobacterales isolates via the Vitek 2 microbial identification system (bioMérieux, Athens, GA) or by the MicroScan WalkAway system (Siemens Healthcare Diagnostics, Tarrytown, NY). Screening for production of extended-spectrum β-lactamases involved testing with one or more of the following agents: ceftazidime, ceftriaxone, cefotaxime, aztreonam, or cefpodoxime, based on guidance from the Clinical and Laboratory Standards Institute (CLSI) (Clinical and Laboratory Standards Institute, 2023). ESBL production was confirmed on the automated instruments or by disk diffusion assays (BBL; Becton, Dickinson and Company, Sparks, MD) or by the measurement of minimum inhibitory concentrations (MICs) of cefotaxime or ceftazidime in the presence and absence of clavulanic acid. The ESBL phenotype is confirmed based on CLSI recommendations during the time period in which the isolates were analyzed (Clinical and Laboratory Standards Institute, 2023).
Isolates that displayed the carbapenemase phenotype were non-susceptible to all extended-spectrum cephalosporins (cefotaxime, ceftazidime, or ceftriaxone) and resistant to one or more carbapenem (imipenem, meropenem, doripenem, or ertapenem) as per Centers for Disease Control and Prevention (CDC) criteria (Centers for Disease Control and Prevention, 2015). Carbapenemase production was confirmed by phenotypic testing or molecular assays, based on guidance available during the time period in which the isolates were analyzed, as appropriate (Birgy et al., 2012; Centers for Disease Control and Prevention, 2019).
Determination of mechanisms of β-lactam resistance
Genomic DNA was purified from Enterobacterales isolates confirmed with an ESBL or carbapenemase-producing phenotype (DNeasy blood and tissue kit, Qiagen, Inc. Valencia, CA). DNA Microarray (Check-MDR CT101 and CT103XL; Check-Points, Wageningen, Netherlands) or polymerase chain reaction (PCR) was utilized for confirmation of the presence of bla genes in Enterobacterales isolates as previously described by our laboratory and according to the manufacturer’s protocol (Logan et al., 2016; Powell et al., 2017).
Determination of mechanisms of fluoroquinolone resistance
To investigate the presence of FQR determinants among β-lactamase-producing Enterobacterales that were both phenotypically susceptible and resistant to fluoroquinolones, we analyzed the quinolone resistance-determining region (QRDR) located on the bacterial chromosome and assessed for plasmid-mediated mechanisms of resistance using CLSI standards (Clinical and Laboratory Standards Institute, 2023). Briefly, assays for mutations in gyrA and parC genes of the QRDR (in E. coli, Klebsiella spp., and Enterobacter spp.) and for plasmid-mediated resistance mechanisms were performed by PCR and/or deoxyribonucleic acid (DNA) sequencing (Robicsek et al., 2006). Extraction of genomic DNA followed by amplification and sequencing were performed using primers and methods as previously described (Cano et al., 2009; Endimiani et al., 2009; Hujer et al., 2009). Specific PMFQR genes for analysis included aac(6’)-Ib-cr, qepA, qnrA, qnrB, qnrC, qnrD, qnrS, oqxA, and oqxB and represented transmissible elements previously reported in Enterobacterales (Chen et al., 2012; Jacoby, 2017; Logan et al., 2019b).
Nomenclature and characterization of Enterobacterales isolates
E. coli was classified into one of four phylogenetic groups (A, B1, B2, and D) using a well-established multiplex PCR-based technique (Bingen-Bidois et al., 2002). Multi-locus sequence typing (MLST) was performed for PMFQR Ent isolates using the Pasteur scheme [Pasteur website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/)] to identify sequence types of ESBL and carbapenemase-producing strains of E. coli and K. pneumoniae, and the Oxford scheme [PubMLST website (https://pubmlst.org/ecloacae/)] was likewise used on Enterobacter cloacae isolates as previously described by our laboratory and others (Diancourt et al., 2005; Jaureguy et al., 2008; Logan et al., 2016; Jolley et al., 2018).
Whole genome sequencing on FQS Enterobacterales strains
For PMFQS Ent strains, DNA was extracted using the MasterPure Gram Positive DNA purification kit following the manufacturer’s instructions (Epicentre, Madison, WI). Libraries were prepared using the Oxford Nanopore Technology’s Rapid Barcoding Kit and whole genome sequencing (WGS) was performed in an ONT MinION. The genome was de novo assembled with Flye 2.9, polished with Medaka 1.6 annotated using BV-BRC (Bacterial and viral bioinformatics resource center) and confirmed with BLAST (Basic Local Alignment Search Tool, NCBI). Resistome and plasmid types were determined using ResFinder 4.0 and Plasmidfinder 2.1, respectively, from the Center for Genomic Epidemiology (https://www.genomicepidemiology.org/services/).
Analytic study design
We used a retrospective case–control study design to assess factors associated with infection due to β-lactam-resistant Enterobacterales, which were phenotypically susceptible to fluoroquinolones yet possessed an FQR gene. We selected as cases those who had infections by FQ-susceptible, β-lactam-resistant Enterobacterales harboring a non- or low-level expression of PMFQR genes (PMFQS Ent). We chose to analyze mechanisms of FQR in detail because this antibiotic class is not commonly used in children, yet 50% of the isolates were fluoroquinolone resistant.
Children with β-lactam-resistant Enterobacterales infections that were phenotypically resistant to fluoroquinolones and in which we had detected an FQR gene (PMFQR Ent) were selected as controls. Specifically, the control group included children with infections that were resistant to extended-spectrum cephalosporins, carbapenem, and fluoroquinolone antibiotics to understand differences between children who had Enterobacterales infections that were phenotypically susceptible to fluoroquinolones and those who had fluoroquinolone phenotypically resistant infections.
Only pediatric patients with clinical infections were included, as determined by using standard criteria defined by the CDC National Healthcare Safety Network and by a study investigator case review (Bingen-Bidois et al., 2002).
Covariates
Multiple variables were analyzed as potential factors associated with PMFQS Ent infection based on known associations for acquisition in adults and children including (1) demographics (age, gender, and race/ethnicity); (2) comorbid conditions (as defined by ICD-9/ICD-10 codes); (3) recent healthcare exposures in both inpatient and outpatient settings, including hospitalization and/or procedures in the previous 30 days; (4) all recent antibiotic exposures in the 40 days prior to culture; and (5) presence, number, and type of invasive medical devices.
Statistical analysis
First, case and control groups were examined for differences using parametric or non-parametric tests as appropriate for categorical and continuous variables, where p ≤ 0.05 was considered statistically significant unless otherwise stated. Next, variables with p < 0.1 on bivariate analysis were incorporated in the multivariable analysis. We then used stepwise multiple logistic regression to assess the multivariable relationship between the covariates and the groups. The final multivariable logistic regression model included the parsimonious model with significant covariates (p < 0.05) from the stepwise selection process, with PMFQS Ent infection as the dichotomous outcome variable. During the model building stages, we did not find evidence of significant confounding; therefore, the simplest model was chosen based on a relatively small sample size and the effect of variables in the model. All analyses were performed in SAS 9.4 (SAS Institute, Cary, NC, USA).
Results
Composition of fluoroquinolone resistance genes in Enterobacterales
We analyzed 170 bla-producing Enterobacterales isolates from Hospitals A, B, and C for the presence of FQR genes (Table 1). Of 170 Enterobacterales isolates, 85 (50%) were FQR and 85 (50%) were FQS. Of the 85 FQS isolates, 23 (27%) had FQR genes detected (PMFQS cases); and of the 85 FQR isolates, 53 (62%) had FQR genes detected (PMFQR controls).
Among the subgroups with plasmid-mediated FQR genes detected by PCR, the median age of 23 children with FQS Enterobacterales and the 53 children with FQR Enterobacterales infections was 4.3 and 6.2 years, respectively (p = NS). The most common source type was urine, accounting for 52.2% of PMFQS and 69.8% of PMFQR infection sources, followed by the respiratory tract in 34.8% and 13.2% of PMFQS cases and PMFQR controls, respectively. The predominant organisms among PMFQS cases were Klebsiella spp. (56.5%) and the most common bla genotype was blaSHV ESBL in 43.5% of cases, whereas in PMFQR controls, the predominant organisms were E. coli (75.5%, p < 0.001), and the most common bla genotype found associated was blaCTX-M-1-group ESBL in 79.2% of controls. The most common quinolone resistance genes among PMFQS Ent and PMFQR Ent isolates were oqxAB and aac(6’)-Ib-cr in 56.5% and 83.0% of isolates, respectively (p < 0.001). Chromosomal resistance mutations in the QRDR region (gyrA and parC) were found in 8.7% of PMFQS Ent and 87.8% of PMFQR Ent isolates. Multidrug resistance (displayed resistance to three or more antibiotic classes) among PMFQS Ent and PMFQR Ent was common, representing 73.9% and 98.1% of isolates, respectively.
Whole genome sequencing analysis of PMFQS Enterobacterales Isolates
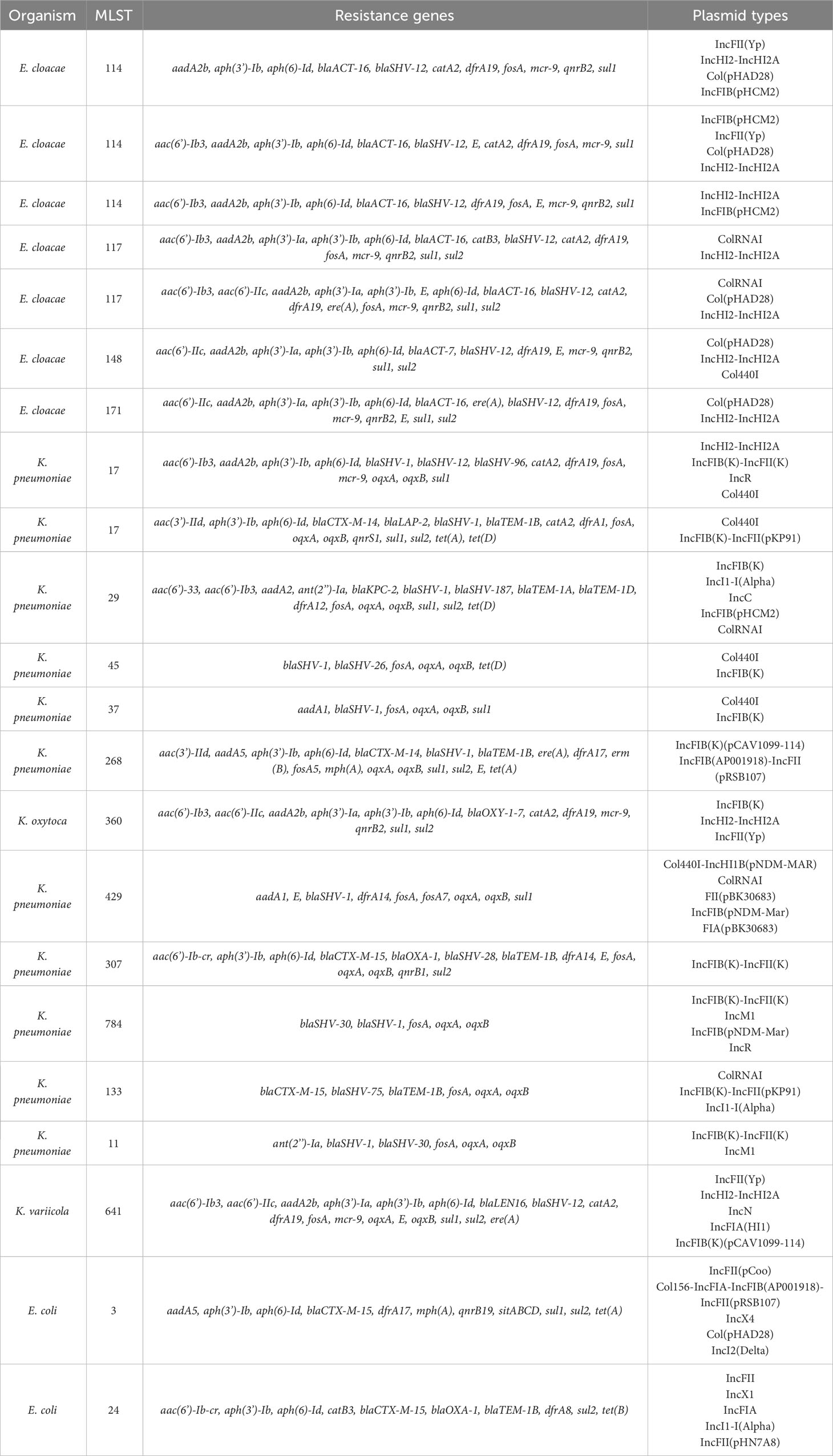
WGS analysis was performed on 22 of 23 available PMFQS Ent isolates to further analyze genetic determinants of antibiotic resistance (resistome) and the mobile genetic elements associated with propagation of drug resistance. We sought to understand if PMFQS Ent isolates that have non- or low-level gene expression of FQR genes may also display “silent” resistance determinants to other antibiotic classes. Multiple plasmid and strain types were found to be associated with drug resistance among PMFQS Ent isolates (Table 2).
Multi-locus sequence typing of the 22 E. coli, Klebsiella spp., and E. cloacae revealed diverse strain types. The most common plasmid replicons present in strains were IncFI and IncFII. An unexpected finding in the resistome was the presence of mcr-9 in 10 (43.5%), a gene associated with transferrable polymyxin resistance. The mcr-9 gene was found in E. cloacae, K. pneumoniae, and K. oxytoca and carried on IncHI2 plasmids. Figure 1 reveals a close relationship between mcr-9 harboring plasmids in different strains, based on protein sequence identity, with the K. oxytoca strain exhibiting the most significant genetic divergence. Figure 2 highlights the complexity of the mcr-9 harboring plasmid and the associated genetic environment.
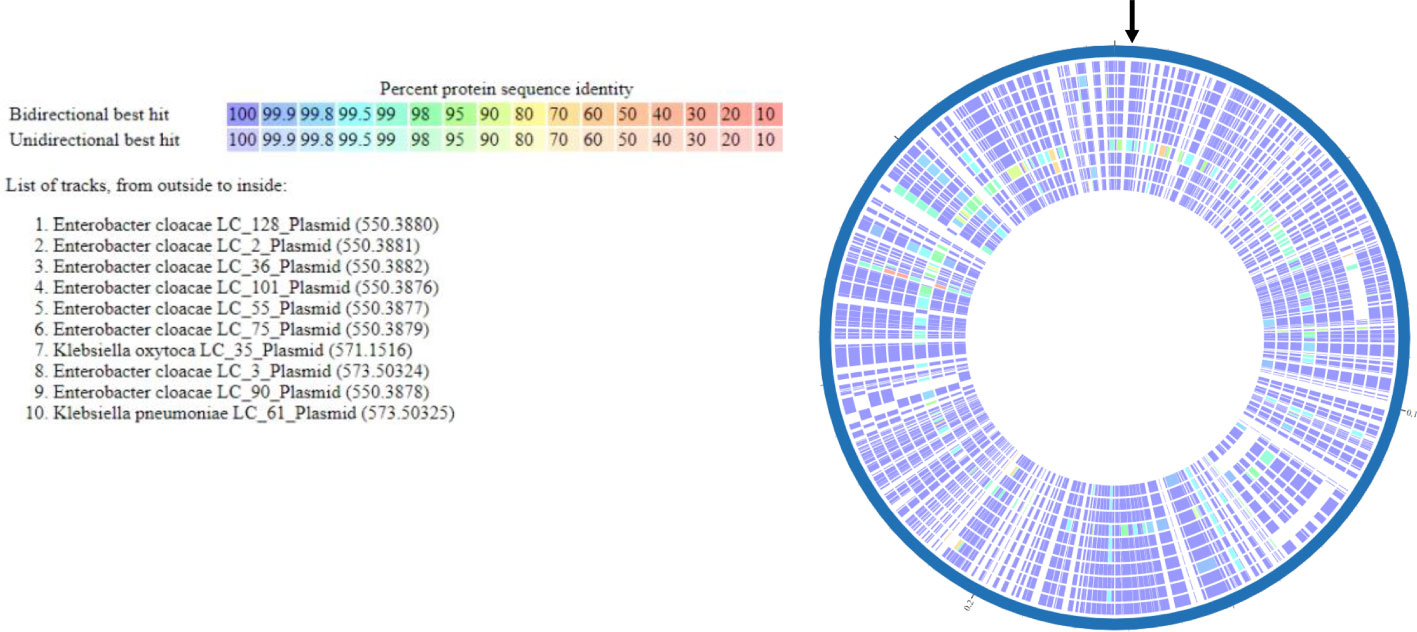
Figure 1 mcr-9 plasmid proteome comparison among PMFQS Enterobacterales strains generated by the BV-BRC Bacterial Proteome Comparison Service. The location of mcr-9 is indicated by the black arrow.
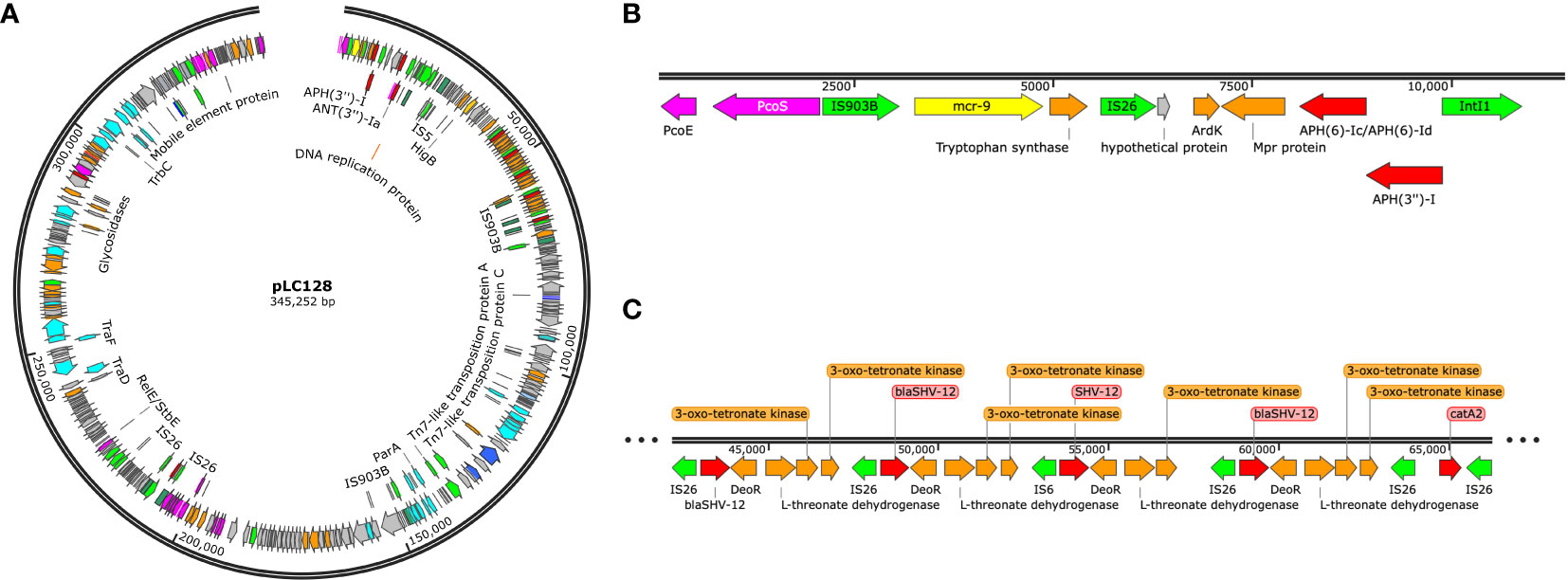
Figure 2 (A) mcr-9 containing plasmid found in Enterobacter cloacae strain LC128, visualized using Snapgene. (B) mcr-9 genetic environment. (C) blaSHV-12 gene amplification.
Analysis of factors associated with PMFQS Enterobacterales infections in children
The 23 cases of PMFQS Ent infection were compared to 53 controls with PMFQR Ent infections. Significant factors positively associated with PMFQS Ent infection on bivariate analysis included the following: male gender, infection with Klebsiella spp. or Enterobacter spp., hospital onset infection, location in PICU or NICU settings at the time of diagnosis, comorbid hematologic–oncologic conditions, and having indwelling devices (Table 3). Cases with PMFQS Ent infection were less likely than controls to have infection with E. coli.
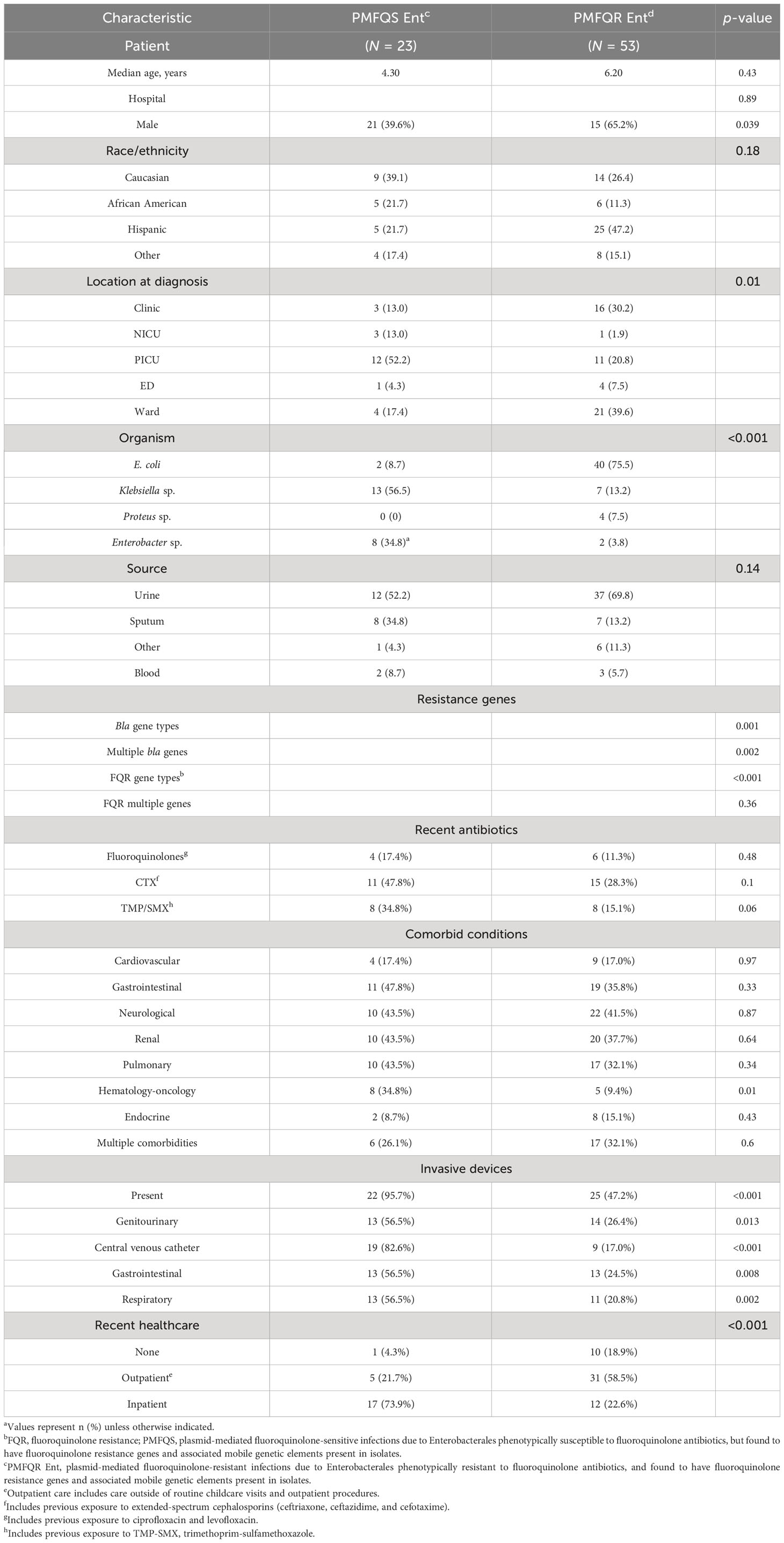
Table 3 Bivariate analysis of demographics and factors associated with PMFQS Enterobacterales infection.
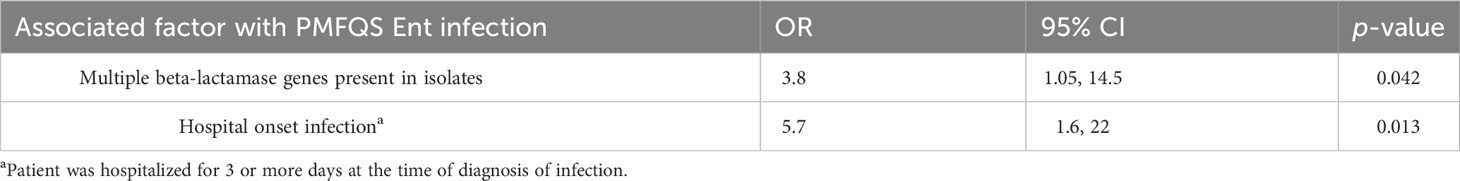
The results of the multivariable regression analysis are shown in Table 4. Cases with PMFQS Ent infection were more likely to have hospital-onset infection, defined as infection that began on or later than day 3 of hospitalization (OR 5.7, 95% CI 1.6, 22, p = 0.0013). The presence of invasive devices mediated the effects of healthcare setting in the final model. Cases with PMFQS Ent infection were also more likely than controls to have isolates with multiple β-lactamase genes present (OR 3.8, 95% CI, p = 0.04), which were often associated with the presence of multiple mobile genetic elements (Table 2).
Discussion
In this study, we focused on understanding the genetic basis of FQR in β-lactamase-producing Enterobacterales that were associated with infections in children from multiple centers. We found that among children, there are antibiotic resistance genes on mobile genetic elements that are silently circulating. This non- or low-level expression of resistance is associated with phenotypic susceptibility to antibiotics, yet these resistance genes are transmissible between bacteria, animals, humans, and the environment. Silent dissemination of these plasmids is concerning, especially in a vulnerable population, children. Children are known to be transmitters of many infectious agents, and in the case of MDR Gram-negative bacteria such as the Enterobacterales, children have been found to stay colonized for months to years (Zerr et al., 2014).
Much of the propagation of MDR Enterobacterales is related to the explosion of ESBL-producing bacteria over the past three decades, in particular the ST131 E. coli harboring blaCTX-M and other high-risk clones containing IncFII plasmids with additional genetic structures such as transposons, integrons, and insertion sequences associated with multiple antibiotic resistance gene cassettes (Lukac et al., 2015). These clonal strains are resistant to multiple antibiotics, including the fluoroquinolones, and this resistance has been found to be both chromosomal- and plasmid-mediated (Canton et al., 2012; Medernach and Logan, 2018).
In adults, the rise of fluoroquinolone-resistant Enterobacterales was first recognized in the 1980s, and the increases were associated with widespread fluoroquinolone overuse. This resistance was mainly due to chromosomally based mechanisms, specifically mutations in gyrA and parC genes of the QRDR in E. coli, Klebsiella spp., and Enterobacter spp (Robicsek et al., 2006; Logan et al., 2018; Medernach and Logan, 2018). The most common plasmid-mediated fluoroquinolone resistance (PMFQR) gene in the ST131 E. coli is aac(6’)-Ib-cr, which encodes an aminoglycoside acetyltransferase, whereby the cr mutation additionally confers reduced susceptibility to fluoroquinolones by N-acetylation of its piperazinyl amine (Robicsek et al., 2006).
However, we found that among children, the mechanisms of both β-lactam and FQR are diverse, and there is significant horizontal gene transfer occurring among the Enterobacterales genera (Logan et al., 2016; Logan et al., 2019a; Logan et al., 2019b; Logan et al., 2019c; Logan et al., 2020a). Over 25% of isolates phenotypically susceptible to fluoroquinolones harbored an FQR gene. The associated mobile genetic elements carrying these genes were diverse, with most isolates possessing multiple plasmids and other genetic structures such as transposons and insertion sequences. Paradoxically, fluoroquinolone-sensitive Enterobacterales infections were more often hospital onset, while the fluoroquinolone-resistant infections were more often community onset. Additionally, the fluoroquinolone-susceptible Enterobacterales commonly co-harbored multiple β-lactamase and FQR genes and 74% were MDR. Only 9% of fluoroquinolone-susceptible Enterobacterales had chromosomal mutations in the QRDR, i.e., gyrA and parC (versus 88% of the fluoroquinolone-resistant Enterobacterales). This is not surprising, as typical phenotypic expression of resistance often requires a chromosomal-based mechanism. However, multiple plasmid-mediated genes in Enterobacterales can also be associated with the higher-level expression of resistance, but in our PMFQS cases, 22% of isolates had multiple PMFQR genes that were still found to be susceptible by automated antimicrobial susceptibility systems in the clinical microbiology laboratories, which use CLSI breakpoints (Clinical and Laboratory Standards Institute, 2023).
An unexpected finding was the presence of the mobile colistin resistance (mcr) gene, mcr-9, in 44% of the PMFQS Enterobacterales infection cases. In November 2015, Liu et al. first described transmissible polymyxin resistance in Enterobacterales associated with the plasmid-mediated colistin resistance gene, mcr-1, a member of the phosphoethanolamine transferase enzyme family (Liu et al., 2015). E. coli and K. pneumoniae that possessed mcr-1 were found in colonized food animals, contaminated retail meat, and inpatients in five Chinese provinces (Liu et al., 2015; Logan and Weinstein, 2017). The first reported US case of Enterobacterales harboring the mcr-1 gene was in May of 2016 (Kline et al., 2016), though since this time, others found that mcr-1 was silently disseminating in Enterobacterales in the US prior to this first report (Castanheira et al., 2016).
Based on data from the National Database of Antibiotic Resistant Organisms (NDARO) (https://www.ncbi.nlm.nih.gov/pathogens/antimicrobialresistance/), mcr-9 was first detected in a Salmonella enterica serovar Schwarzengrund recovered from an avian source in the US in 2014, and the first detection in a human clinical isolate was an S. enterica serovar Typhimurium in the US in 2015 (Ling et al., 2020). The majority of the isolates in this current report were infections that occurred in children between 2011 and 2014, suggesting that mcr-9 was silently disseminating many years in Enterobacterales prior to its first description and that children may have been a factor in the rapid global dissemination of these novel transmissible resistance genes. By 2019, mcr-9 was the second most widely disseminated among the MCR-family genes (behind mcr-1), being identified in isolates recovered from humans, animals, and the environment in 40 countries across six continents (Ling et al., 2020).
While most commonly carried by Salmonella species, mcr-9 has now been detected in several genera of Enterobacterales. The suspected origins of mcr-9 are livestock and poultry, raising concern that the sources of human acquisition may be linked to community-based environmental influences that include higher exposure risks in certain communities due to certain foods, animals, livestock, water sources, soil, fertilizer, and vegetation (Silbergeld et al., 2008; Ling et al., 2020). Additionally, some of the FQR genes, for example, oqxA and oqxB, which were the most common FQR genes detected in Klebsiella spp. in our study, are multidrug efflux pumps named for their resistance to olaquindox, which is used as a growth promoter on pig farms (Hansen et al., 2005). Furthermore, we conducted prior pilot studies using metagenomic sequencing and culture-based approaches to analyze whether waterways in Chicago used for limited contact recreation (e.g., kayaking, boating, and fishing) were potential reservoirs of MDR Enterobacterales. We found multiple antibiotic resistance genes among Enterobacterales in surface waters, including mcr-1, which was detected in three of the four evaluated waterways (Logan et al., 2020b).
In conclusion, we found that children with infections due to β-lactamase-producing Enterobacterales often have other antibiotic resistance genes that may have non- or low-level expression resulting in phenotypic susceptibility to antibiotics, yet these genes are transmissible, and the associated mobile genetic elements and strains are able to silently disseminate. Future studies will assess for the presence of β-lactamase genes, FQR genes, and other antibiotic resistance genes, including MCR-family genes in children and healthy populations, in order to further characterize and validate community sources of MDR Enterobacterales, as well as the risk factors associated with colonization and infection in humans. This will be critical to devise the most effective treatment and prevention strategies for these serious infections in our most vulnerable population, children.
Data availability statement
The data presented in the study are deposited in the NCBI Genbank repository, under accession number PRJNA989624.
Ethics statement
The studies involving humans were approved by Ann & Robert H. Lurie Children’s Hospital of Chicago, Rush University Medical Center, Loyola University Medical Center. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because this was a retrospective study of salvage isolates obtained for clinical purposes other than study. The study was approved by the institutional review boards of the three participating institutions and the need for informed consent was waived by all institutions.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work, including the efforts of LL, was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) (K08AI112506), the Marcus Foundation, and by the Emory University School of Medicine MP3 Initiative Award Program. RB is supported by the Department of Veterans Affairs Research and Development under award number I01BX001974, VISN 10 Geriatrics Research, Education and Clinical Center.
Acknowledgments
We thank the microbiology laboratories of the participating institutions for providing isolates for this study. We thank Kendrick Reme of the Logan Laboratory and Pamela Hagen, Ellen Gough, Diane Springer, Jane Stevens, Joyce Houlihan, Cindy Bethel, Kathleen McKinley, Violeta Rekasiu, and Donna Carter of participating institutions for collection, shipping, and cultivation of organisms. We thank the team of curators of the Institut Pasteur MLST and whole-genome MLST databases and NIH GenBank database for curating the data and making them publicly available at http://bigsdb.web.pasteur.fr/ and https://www.ncbi.nlm.nih.gov/genbank/. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bingen-Bidois, M., Clermont, O., Bonacorsi, S., Terki, M., Brahimi, N., Loukil, C., et al. (2002). Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect. Immun. 70 (6), 3216–3226. doi: 10.1128/IAI.70.6.3216-3226.2002
Birgy, A., Bidet, P., Genel, N., Doit, C., Decré, D., Arlet, G., et al. (2012). Phenotypic screening of carbapenemases and associated beta-lactamases in carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 50 (4), 1295–1302. doi: 10.1128/JCM.06131-11
Cano, M. E., Rodriguez-Martinez, J. M., Aguero, J., Pascual, A., Calvo, J., García-Lobo, J. M., et al. (2009). Detection of plasmid-mediated quinolone resistance genes in clinical isolates of Enterobacter spp. Spain J. Clin. Microbiol. 47 (7), 2033–2039. doi: 10.1128/JCM.02229-08
Cantón, R., Coque, T. M. (2006). The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9 (5), 466–475. doi: 10.1016/j.mib.2006.08.011
Canton, R., Gonzalez-Alba, J. M., Galan, J. C. (2012). CTX-M enzymes: Origin and diffusion. Front. Microbiol. 3, 110. doi: 10.3389/fmicb.2012.00110
Castanheira, M., Griffen, M., Deshpande, L., Mendes, R. E., Jones, R., Flamm, R. (2016). Detection of mcr-1 among Escherichia coli Clinical isolates collected worldwide as part of the SENTRY antimicrobial surveillance program in 2014 and 2015. Antimicrob. Agents Chemother. 60 (9), 5623–5624. doi: 10.1128/AAC.01267-16
Centers for Disease Control and Prevention. (2015). CRE toolkit–guidance for control of carbapenem-resistant Enterobacteriaceae (CRE) (Atlanta, GA: CDC).
Centers for Disease Control and Prevention. (2019). Healthcare-Associated Infections: CRE Technical Information. Available at: https://www.cdc.gov/hai/organisms/cre/index.html.
Chen, X., Zhang, W., Pan, W., Yin, J., Pan, Z., Gao, S., et al. (2012). Prevalence of qnr, aac(6’)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and environment. Antimicrob. Agents Chemother. 56 (6), 3423–7. doi: 10.1128/AAC.06191-11
Clinical and Laboratory Standards Institute. (2023). Performance standards for antimicrobial susceptibility testing: M100-ED33. Thirty-third informational supplement (March 2023 update). Available at: https://clsi.org/all-free-resources/.
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A., Brisse, S. (2005). Multilocus sequence typing of klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43 (8), 4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005
Endimiani, A., Hujer, A. M., Perez, F., Bethel, C. R., Hujer, K. M., Kroeger, J., et al. (2009). Characterization of blaKPC-containing klebsiella pneumoniae isolates detected in different institutions in the eastern USA. J. Antimicrob. Chemother. 63 (3), 427–437. doi: 10.1093/jac/dkn547
Hansen, L. H., Sørensen, S. J., Jørgensen, H. S., Jensen, L. B. (2005). The prevalence of the OqxAB multidrug efflux pump amongst olaquindox-resistant Escherichia coli in pigs. Microb. Drug Resist. 11 (4), 378–382. doi: 10.1089/mdr.2005.11.378
Hujer, K. M., Hujer, A. M., Endimiani, A., Thomson, J. M., Adams, M. D., Goglin, K., et al. (2009). Rapid determination of quinolone resistance in Acinetobacter spp. J. Clin. Microbiol. 47 (5), 1436–1442. doi: 10.1128/JCM.02380-08
Jacoby, G. A. (2017). “Plasmid-mediated quinolone resistance,” in Antimicrobial drug resistance (Springer), 265–268. doi: 10.1007/978-3-319-46718-4_17
Jaureguy, F., Landraud, L., Passet, V., Diancourt, L., Frapy, E., Guigon, G., et al. (2008). Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics 9, 560–2164-9-560. doi: 10.1186/1471-2164-9-560
Jolley, K. A., Bray, J. E., Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3, 124. doi: 10.12688/wellcomeopenres.14826.1
Kline, K., Shover, J., Kallen, A., Lonsway, D., Watkins, S., Miller, J. (2016). Investigation of first identified mcr-1 gene in an isolate from a US patient—Pennsylvania, 2016. MMWR Morb. Mortal. Wkly. Rep. 65 (36), 977–978. doi: 10.15585/mmwr.mm6536e2
Ling, Z., Yin, W., Shen, Z., Wang, Y., Shen, J., Walsh, T. (2020). Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chem. 75 (11), 3087–3095. doi: 10.1093/jac/dkaa205
Liu, Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al (2015). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 16 (2), 161–8. doi: 10.1016/S1473-3099(15)00424-7
Logan, L. K., Blumer, J., Toltzis, P. (2018). “Antibiotic resistance,” in Feigin and cherry’s textbook of pediatric infectious diseases, 8th ed. Eds. Cherry, J., Demmler-Harrison, G. J., Kaplan, S. L., Steinbach, W. J., Hotez, P. J. (Elsevier). Chapter 234.
Logan, L. K., Braykov, N. P., Weinstein, R. A., Laxminarayan, R. (2014). Extended-spectrum β-lactamase–producing and third-generation cephalosporin-resistant Enterobacteriaceae in children: Trends in the United States, 1999–2011. J. Pediatr. Infect. Dis. Society. 3 (4), 320–328. doi: 10.1093/jpids/piu010
Logan, L. K., Hujer, A. M., Marshall, S. H., Domitrovic, T. N., Rudin, S. D., Zheng, X., et al. (2016). Analysis of beta-lactamase resistance determinants in Enterobacteriaceae from Chicago children: A multicenter survey. Antimicrob. Agents Chemother. 60 (6), 3462–3469. doi: 10.1128/AAC.00098-16
Logan, L. K., Medernach, R. L., Domitrovic, T. N., Rispens, J. R., Hujer, A. M., Qureshi, N. K., et al. (2019a). The clinical and molecular epidemiology of CTX-M-9 group producing Enterobacteriaceae infections in children. Infect. Dis. Ther. 8 (2), 243–254. doi: 10.1007/s40121-019-0237-2
Logan, L. K., Medernach, R. L., Rispens, J. R., Marshall, S. H., Hujer, A. M., Domitrovic, T. N., et al. (2019b). Community origins and regional differences highlight risk of plasmid-mediated fluoroquinolone resistant Enterobacteriaceae infections in children. Pediatr. Infect. Dis. J. 38 (6), 595–599. doi: 10.1097/INF.0000000000002205
Logan, L. K., Nguyen, D. C., Scaggs-Huang, F. A., Qureshi, N. K., Charnot-Katsikas, A., Bartlett, A. H., et al. (2019c). A multi-centered case-case-control study of factors associated with klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae (KPC-CRE) infections in children and young adults. Pediatr. Infect. Dis. J. 38 (5), 490–495. doi: 10.1097/INF.0000000000002176
Logan, L. K., Renschler, J. P., Gandra, S., Weinstein, R. A., Laxminarayan, R., Centers for Disease Control, et al. (2015). Carbapenem-resistant enterobacteriaceae in children, United States, 1999–2012. Emerg. Infect. Dis. 21 (11), 2014–2021. doi: 10.3201/eid2111.150548
Logan, L. K., Rispens, J. R., Medernach, R. L., Domitrovic, T. N., Hujer, A. M., Marshall, S. H., et al. (2020a). A multicentered study of the clinical and molecular epidemiology of TEM-and SHV-type extended-spectrum beta-lactamase producing Enterobacterales infections in children. Pediatr. Infect. Dis. J. 40 (1), 39–43. doi: 10.1097/INF.0000000000002916
Logan, L. K., Weinstein, R. A. (2017). The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 215 (suppl_1), S28–S36. doi: 10.1093/infdis/jiw282
Logan, L. K., Zhang, L., Green, S., Dorevitch, S., Arango-Argoty, G. A., Reme, K., et al. (2020b). A pilot study of waterways as reservoirs of multi-drug resistant Enterobacteriaceae (MDR Ent) in a high-risk region for community-acquired MDR-Ent infection in children. Antimicrob. Agents Chemother. 64 (4), e02310–e02319. doi: 10.1128/AAC.02310-19
Lukac, P. J., Bonomo, R. A., Logan, L. K. (2015). Extended-spectrum beta-lactamase-producing Enterobacteriaceae in children: Old foe, emerging threat. Clin. Infect. Dis. 60 (9), 1389–1397. doi: 10.1093/cid/civ020
Mathers, A. J., Peirano, G., Pitout, J. D. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28 (3), 565–591. doi: 10.1128/CMR.00116-14
Medernach, R. L., Logan, L. K. (2018). The growing threat of antibiotic resistance in children. Infect. Dis. Clin. North Am. 32 (1), 1–17. doi: 10.1016/j.idc.2017.11.001
Pitout, J. D., Peirano, G., Chen, L., DeVinney, R., Matsumura, Y. (2022). Escherichia coli ST1193: Following in the footsteps of E. coli ST131. J. Antimicrob. Chem. 66 (7), e00511–e00522. doi: 10.1128/aac.00511-22
Powell, E. A., Haslam, D., Mortensen, J. E. (2017). Performance of the checkpoints check-MDR CT103XL assay utilizing the CDC/FDA antimicrobial resistance isolate bank. Diagn. Microbiol. Infect. Dis. 88 (3), 219–221. doi: 10.1016/j.diagmicrobio.2017.04.011
Price, L. B., Johnson, J. R., Aziz, M., Clabots, C., Johnston, B., Tchesnokova, V., et al. (2013). The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-rx. MBio 4 (6), e00377–e00313. doi: 10.1128/mBio.00377-13
Robicsek, A., Jacoby, G. A., Hooper, D. C. (2006). The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6 (10), 629–640. doi: 10.1016/S1473-3099(06)70599-0
Silbergeld, E. K., Graham, J., Price, L. B. (2008). Industrial food animal production, antimicrobial resistance, and human health. Annu. Rev. Public Health 29, 151–169. doi: 10.1146/annurev.publhealth.29.020907.090904
Tchesnokova, V. L., Rechkina, E., Larson, L., Ferrier, K., Weaver, J. L., Schroeder, D. W., et al. (2018). Rapid and extensive expansion in the United States of a new multidrug-resistant Escherichia coli clonal group, sequence type 1193. Clin. Infect. Dis. 68 (2), 334–337. doi: 10.1093/cid/ciy525
Keywords: epidemiology, gram-negative bacteria, Enterobacterales infections, fluoroquinolone resistance, beta-lactamases, children, antibiotic resistance, mcr-9
Citation: Logan LK, Coy LR, Pitstick CE, Marshall SH, Medernach RL, Domitrovic TN, Konda S, Qureshi NK, Hujer AM, Zheng X, Rudin SD, Weinstein RA and Bonomo RA (2023) The role of the plasmid-mediated fluoroquinolone resistance genes as resistance mechanisms in pediatric infections due to Enterobacterales. Front. Cell. Infect. Microbiol. 13:1249505. doi: 10.3389/fcimb.2023.1249505
Received: 28 June 2023; Accepted: 30 August 2023;
Published: 09 October 2023.
Edited by:
Christoph Gabler, Freie Universität Berlin, GermanyReviewed by:
Sophia Vourli, University General Hospital Attikon, GreeceNubwa Medugu, Nile University of Nigeria, Nigeria
Copyright © 2023 Logan, Coy, Pitstick, Marshall, Medernach, Domitrovic, Konda, Qureshi, Hujer, Zheng, Rudin, Weinstein and Bonomo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Latania K. Logan, LKLogan@emory.edu
†Present addresses: Claire Pitstick, Private Practice Pediatrics, Chicago, IL, United States
Xiaotian Zheng, Department of Pathology and Laboratory Medicine, Akron Children’s Hospital, Akron, OH, United States
Sreenivas Konda, Department of Population and Precision Health, University of Chicago, Chicago, IL, United States
T. Nicholas Domitrovic, University of Toledo College of Medicine, Toledo, OH, United States
‡These authors have contributed equally to this work
 Latania K. Logan
Latania K. Logan Laura Rojas Coy
Laura Rojas Coy Claire E. Pitstick3†‡
Claire E. Pitstick3†‡  Steven H. Marshall
Steven H. Marshall Rachel L. Medernach
Rachel L. Medernach T. Nicholas Domitrovic
T. Nicholas Domitrovic Sreenivas Konda
Sreenivas Konda Andrea M. Hujer
Andrea M. Hujer