Abstract
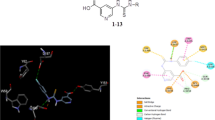
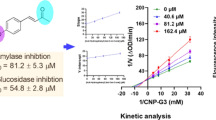
We aimed to identify and evaluate natural organosulfur compounds (OSCs) from the genus Allium as potent dual inhibitors of α-amylase and α-glucosidase via a set of molecular simulations and in-vitro analysis. We reported that all OSCs fall under the acceptable scores of Lipinski’s rules and desirable ADME parameters. In-silico screening revealed that OSCs strongly interacted with α-amylase and α-glucosidase crystal structures. Our in-vitro studies with four OSCs having best ΔG-scores demonstrated that alliin, S-allyl-L-cysteine (SAC), N-acetylcysteine (NAC), and S-ethyl-L-cysteine (SEC) exhibit a marked in-vitro inhibition of α-amylase activity, whereas, γ-glutamylcysteine (GGC), SEC, alliin, and SAC potentially inhibited the in-vitro α-glucosidase activity. Our enzyme kinetics studies depicted that the selected OSCs competitively inhibited the α-amylase and α-glucosidase activity. In conclusion, this initial in-silico and the in-vitro report demonstrates the drug-likeness and ADME indices of OSCs and identifies alliin, SAC, NAC, SEC, and GGC as potential dual inhibitors of α-amylase and α-glucosidase.











Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published paper.
Change history
22 October 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00044-021-02807-5
References
Nabi R, Alvi SS, Saeed M, Ahmad S, Khan MS. Glycation and HMG-CoA reductase inhibitors: implication in diabetes and associated complications. Curr Diabetes Rev. 2019;15:213–23. https://doi.org/10.2174/1573399814666180924113442
Nabi R, Alvi SS, Shah A, Chaturvedi CP, Faisal M, Alatar AA, et al. Ezetimibe attenuates experimental diabetes and renal pathologies via targeting the advanced glycation, oxidative stress and AGE-RAGE signalling in rats. Arch Physiol Biochem. 2021. https://doi.org/10.1080/13813455.2021.1874996. [Online fisrt article]
Standl E, Khunti K, Hansen TB, Schnell O. The global epidemics of diabetes in the 21st century: current situation and perspectives. Eur J Prev Cardiol. 2019;26:7–14. https://doi.org/10.1177/2047487319881021
Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N, et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pr. 2019;157:107843 https://doi.org/10.1016/j.diabres.2019.107843
Hashim A, Alvi SS, Ansari IA, Khan MS. Phyllanthus virgatus forst extract and it’s partially purified fraction ameliorates oxidative stress and retino-nephropathic architecture in streptozotocin-induced diabetic rats. Pak J Pharm Sci. 2019;32:2697–708. https://doi.org/10.36721/PJPS.2019.32.6.REG.2697-2708.1
Akhter F, Alvi SS, Ahmad P, Iqbal D, Alshehri BM, Khan MS. Therapeutic efficacy of Boerhaavia diffusa (Linn.) root methanolic extract in attenuating streptozotocin-induced diabetes, diabetes-linked hyperlipidemia and oxidative-stress in rats. Biomed Res Ther. 2019;6:3293–306. https://doi.org/10.15419/bmrat.v6i7.556
Nabi R, Alvi SS, Shah A, Chaturvedi CP, Iqbal D, Ahmad S, et al. Modulatory role of HMG-CoA reductase inhibitors and ezetimibe on LDL-AGEs-induced ROS generation and RAGE-associated signalling in HEK-293 Cells. Life Sci. 2019;235:116823 https://doi.org/10.1016/j.lfs.2019.116823
Nabi R, Alvi SS, Khan RH, Ahmad S, Ahmad S, Khan MS. Antiglycation study of HMG-R inhibitors and tocotrienol against glycated BSA and LDL: A comparative study. Int J Biol Macromol. 2018;116:983–92. https://doi.org/10.1016/j.ijbiomac.2018.05.115
Nabi R, Alvi SS, Shah MS, Ahmad S, Faisal M, Alatar AA, et al. A biochemical & biophysical study on in-vitro anti-glycating potential of iridin against D-Ribose modified BSA. Arch Biochem Biophys. 2020;686:108373 https://doi.org/10.1016/j.abb.2020.108373
Nabi R, Alvi SS, Alouffi S, Khan S, Ahmad A, Khan M, et al. Amelioration of neuropilin-1 and rage/matrix metalloproteinase-2 pathway-induced renal injury in diabetic rats by rosuvastatin. Arch Biol Sci. 2021;73:265–78. https://doi.org/10.2298/abs210316021n
Bhatia A, Singh B, Arora R, Arora S. In vitro evaluation of the α-glucosidase inhibitory potential of methanolic extracts of traditionally used antidiabetic plants. BMC Complement Alter Med. 2019;19:74 https://doi.org/10.1186/s12906-019-2482-z
Hashim A, Khan MS, Khan MS, Baig MH, Ahmad S. Antioxidant and αmylase inhibitory property of phyllanthus virgatus L.: An in vitro and molecular interaction study. Biomed Res Int. 2013;2013:729393 https://doi.org/10.1155/2013/729393
Hess A, Kress S, Rakete S, Muench G, Kornhuber J, Pischetsrieder M, et al. Influence of the fat/carbohydrate component of snack food on energy intake pattern and reinforcing properties in rodents. Behav Brain Res. 2019;364:328–33. https://doi.org/10.1016/j.bbr.2019.02.041
Chaudhury A, Duvoor C, Reddy Dendi VS, Kraleti S, Chada A, Ravilla R, et al. Clinical Review of Antidiabetic Drugs: implications for Type 2 Diabetes Mellitus Management. Front Endocrinol (Lausanne). 2017;8:6 https://doi.org/10.3389/fendo.2017.00006
Alvi SS, Ahmad P, Ishrat M, Iqbal D, Khan MS. Secondary Metabolites from Rosemary (Rosmarinus officinalis L.): Structure, Biochemistry and Therapeutic Implications Against Neurodegenerative Diseases. In: Swamy M, Akhtar M, editors. Nat Bio-active Compd. Singapore: Springer; 2019. p. 1–24. 10.1007/978-981-13-7205-6_1
Ahmad P, Alvi SS, Iqbal D, Khan MS. Insights into pharmacological mechanisms of polydatin in targeting risk factors-mediated atherosclerosis. Life Sci. 2020;254:117756 https://doi.org/10.1016/j.lfs.2020.117756
Kim S, Lee S, Shin D, Yoo M. Change in organosulfur compounds in onion (Allium cepa L.) during heat treatment. Food Sci Biotechnol. 2016;25:115–9. https://doi.org/10.1007/s10068-016-0017-7
Chekki R, Najjaa H. Detection of Organo-Sulphur Volatiles in Allium sativum by Factorial Design. Nat Prod Chem Res. 2016;4:1000211 https://doi.org/10.4172/2329-6836.1000211
Ahmad P, Alvi SS, Khan MS. Functioning of organosulfur compounds from garlic (allium sativum linn) in targeting risk factor-mediated atherosclerosis: a cross talk between alternative and modern medicine. In: Natural Bio-active Compounds: Volume 1: Production and Applications. Springer, Singapore; 2019; p. 561–85. https://doi.org/10.1007/978-981-13-7154-7_20.
Shang A, Cao SY, Xu XY, Gan RY, Tang GY, Corke H, et al. Bioactive compounds and biological functions of garlic (allium sativum L.). Foods. 2019;8:246 https://doi.org/10.3390/foods8070246
De Greef D, Barton EM, Sandberg EN, Croley CR, Pumarol J, Wong TL, et al. Anticancer potential of garlic and its bioactive constituents: a systematic and comprehensive review. Semin Cancer Biol. 2021;73:219–64. https://doi.org/10.1016/j.semcancer.2020.11.020
Batiha GES, Beshbishy AM, Wasef LG, Elewa YHA, Al-Sagan AA, El-Hack MEA, et al. Chemical constituents and pharmacological activities of garlic (Allium sativum L.): a review. Nutrients. 2020;12:872 https://doi.org/10.3390/nu12030872
Ruhee RT, Roberts LA, Ma S, Suzuki K. Organosulfur Compounds: a Review of Their Anti-inflammatory Effects in Human Health. Front Nutr. 2020;7:64 https://doi.org/10.3389/fnut.2020.00064
Schneider P, Walters WP, Plowright AT, Sieroka N, Listgarten J, Goodnow RA, et al. Rethinking drug design in the artificial intelligence era. Nat Rev Drug Discov. 2020;19:353–64. https://doi.org/10.1038/s41573-019-0050-3
Hessler G, Baringhaus K-H. Artificial Intelligence in Drug Design. Molecules. 2018;23:2520 https://doi.org/10.3390/molecules23102520
Pajouhesh H, Lenz GR. Medicinal chemical properties of successful central nervous system drugs. NeuroRx. 2005;2:541–53. https://doi.org/10.1602/neurorx.2.4.541
Hitchcock SA, Pennington LD. Structure-brain exposure relationships. J Med Chem. 2006;49:7559–83. https://doi.org/10.1021/jm060642i
Prasanna S, Doerksen R. Topological Polar Surface Area: A Useful Descriptor in 2D-QSAR. Curr Med Chem. 2008;16:21–41. https://doi.org/10.2174/092986709787002817
Ma XL, Chen C, Yang J. Predictive model of blood-brain barrier penetration of organic compounds. Acta Pharm Sin. 2005;26:500–12. https://doi.org/10.1111/j.1745-7254.2005.00068.x
Leão RP, Cruz JV, da Costa GV, Cruz JN, Ferreira EFB, Silva RC, et al. Identification of new rofecoxib-based cyclooxygenase-2 inhibitors: a bioinformatics approach. Pharmaceuticals. 2020;13:209 https://doi.org/10.3390/ph13090209
Bittermann K, Goss K-U. Predicting apparent passive permeability of Caco-2 and MDCK cell-monolayers: a mechanistic model. PLoS ONE. 2017;12:e0190319 https://doi.org/10.1371/journal.pone.0190319
Volpe DA. Variability in caco-2 and MDCK cell-based intestinal permeability assays. J Pharm Sci. 2008;97:712–25. https://doi.org/10.1002/jps.21010
Yamashita S, Furubayashi T, Kataoka M, Sakane T, Sezaki H, Tokuda H. Optimized conditions for prediction of intestinal drug permeability using Caco-2 cells. Eur J Pharm Sci. 2000;10:195–204. https://doi.org/10.1016/S0928-0987(00)00076-2
Lundborg M, Wennberg CL, Narangifard A, Lindahl E, Norlén L. Predicting drug permeability through skin using molecular dynamics simulation. J Control Release. 2018;283:269–79. https://doi.org/10.1016/j.jconrel.2018.05.026
Chen C-P, Chen C-C, Huang C-W, Chang Y-C. Evaluating Molecular Properties Involved in Transport of Small Molecules in Stratum Corneum: a Quantitative Structure-Activity Relationship for Skin Permeability. Molecules. 2018;23:911 https://doi.org/10.3390/molecules23040911
Roberts JA, Pea F, Lipman J. The clinical relevance of plasma protein binding changes. Clin Pharmacokinet. 2013;52:1–8. https://doi.org/10.1007/s40262-012-0018-5
Gurevich KG. Effect of blood protein concentrations on drug-dosing regimes: practical guidance. Theor Biol Med Model. 2013;10:20 https://doi.org/10.1186/1742-4682-10-20
Zhao YH, Le J, Abraham MH, Hersey A, Eddershaw PJ, Luscombe CN, et al. Evaluation of human intestinal absorption data and subsequent derivation of a quantitative structure - Activity relationship (QSAR) with the Abraham descriptors. J Pharm Sci. 2001;90:749–84. https://doi.org/10.1002/jps.1031
Alvi SS, Ansari IA, Khan MS. Pleiotropic role of lycopene in protecting various risk factors mediated atherosclerosis. Ann Phytomedicine. 2015;4:54–60.
Wang B, Yang LP, Zhang XZ, Huang SQ, Bartlam M, Zhou SF. New insights into the structural characteristics and functional relevance of the human cytochrome P450 2D6 enzyme. Drug Metab Rev 2009;41:573–643. https://doi.org/10.1080/03602530903118729
Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–41. https://doi.org/10.1016/j.pharmthera.2012.12.007
Teh LK, Bertilsson L. Pharmacogenomics of CYP2D6: Molecular genetics, interethnic differences and clinical importance. Drug Metab Pharmacokinet. 2012;27:55–67. https://doi.org/10.2133/dmpk.DMPK-11-RV-121
Walko CM, McLeod H. Use of CYP2D6 genotyping in practice: Tamoxifen dose adjustment. Pharmacogenomics. 2012;13:691–7. https://doi.org/10.2217/pgs.12.27
Neves Cruz J, Santana De Oliveira M, Gomes Silva S, Pedro Da Silva Souza Filho A, Santiago Pereira D, Lima E Lima AH, et al. Insight into the Interaction Mechanism of Nicotine, NNK, and NNN with Cytochrome P450 2A13 Based on Molecular Dynamics Simulation. J Chem Inf Model. 2020;60:766–76. https://doi.org/10.1021/acs.jcim.9b00741
Meena SN, Kumar U, Naik MM, Ghadi SC, Tilve SG. α-Glucosidase inhibition activity and in silico study of 2-(benzo[d][1,3]dioxol-5-yl)-4H-chromen-4-one, a synthetic derivative of flavone. Bioorg Med Chem. 2019;27:2340–4. https://doi.org/10.1016/j.bmc.2018.12.021
Mugaranja KP, Kulal A. Alpha glucosidase inhibition activity of phenolic fraction from Simarouba glauca: An in-vitro, in-silico and kinetic study. Heliyon. 2020;6:e04392 https://doi.org/10.1016/j.heliyon.2020.e04392
Dubey S, Ganeshpurkar A, Ganeshpurkar A, Bansal D, Dubey N. Glycolytic enzyme inhibitory and antiglycation potential of rutin. Futur J Pharm Sci. 2017;3:158–62. https://doi.org/10.1016/j.fjps.2017.05.005
Alvi SS, Iqbal D, Ahmad S, Khan MS. Molecular rationale delineating the role of lycopene as a potent HMG-CoA reductase inhibitor: in vitro and in silico study. Nat Prod Res. 2016;30:2111–4. https://doi.org/10.1080/14786419.2015.1108977
Arvindekar A, Ghadyale V, Takalikar S, Haldavnekar V. Effective control of postprandial glucose level through inhibition of intestinal alpha glucosidase by Cymbopogon martinii (Roxb.). Evid-based Complement Alter Med. 2012;2012:372909 https://doi.org/10.1155/2012/372909
Zhang H, Wang G, Beta T, Dong J. Inhibitory properties of aqueous ethanol extracts of propolis on alpha-glucosidase. Evid-based Complement Alter Med. 2015;2015:587383 https://doi.org/10.1155/2015/587383
Sadeghi M, Moradi M, Madanchi H, Johari B. In silico study of garlic (Allium sativum L.)-derived compounds molecular interactions with α-glucosidase. Silico Pharm. 2021;9:11 https://doi.org/10.1007/s40203-020-00072-9
Hyeong-U S, Eun-Kyeong Y, Chi-Yeol Y, Chul-Hong P, Myung-Ae B, Tae-Ho K, et al. Effects of synergistic inhibition on α-glucosidase by phytoalexins in soybeans. Biomolecules. 2019;9:828 https://doi.org/10.3390/biom9120828
Ge J, Wang Z, Cheng Y, Ren J, Wu B, Li W, et al. Computational study of novel natural inhibitors targeting aminopeptidase N(CD13). Aging (Albany NY). 2020;12:8523–35. https://doi.org/10.18632/aging.103155
Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717 https://doi.org/10.1038/srep42717
Alvi SS, Ansari IA, Khan I, Iqbal J, Khan MS. Potential role of lycopene in targeting proprotein convertase subtilisin/kexin type-9 to combat hypercholesterolemia. Free Radic Biol Med. 2017;108:394–403. https://doi.org/10.1016/j.freeradbiomed.2017.04.012
Jiménez J, Doerr S, Martínez-Rosell G, Rose AS, De Fabritiis G. DeepSite: Protein-binding site predictor using 3D-convolutional neural networks. Bioinformatics. 2017;33:3036–42. https://doi.org/10.1093/bioinformatics/btx350
Wang G, Liu Z, Li M, Li Y, Alvi SS, Ansari IA, et al. Ginkgolide B Mediated Alleviation of Inflammatory Cascades and Altered Lipid Metabolism in HUVECs via Targeting PCSK-9 Expression and Functionality. Biomed Res Int. 2019;2019:7284767 https://doi.org/10.1155/2019/7284767
Alvi SS, Ansari IA, Ahmad MK, Iqbal J, Khan MS. Lycopene amends LPS induced oxidative stress and hypertriglyceridemia via modulating PCSK-9 expression and Apo-CIII mediated lipoprotein lipase activity. Biomed Pharmacother. 2017;96:1082–93. https://doi.org/10.1016/j.biopha.2017.11.116
Du X, Li Y, Xia YL, Ai SM, Liang J, Sang P, et al. Insights into protein–ligand interactions: Mechanisms, models, and methods. Int J Mol Sci. 2016;17:144 https://doi.org/10.3390/ijms17020144
Gurung AB, Ali MA, Lee J, Farah MA, Al-Anazi KM. Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in silico approach. Life Sci. 2020;255:117831 https://doi.org/10.1016/j.lfs.2020.117831
Acknowledgements
The authors are highly indebted to the Department of Health Research (DHR), Ministry of Health & Family Welfare (MoHFW), Govt. of India, for providing the Young Scientist Grant (YSS Project No. 12014/13/2019-HR) for the smooth conduction of this work. The authors are also thankful to Integral University for providing laboratory for the smooth completion of this work. This paper has Integral University manuscript communication no. IU/R&D/2021-MCN0001155.
Author information
Authors and Affiliations
Contributions
Author contributionsInterventional hypothesis: MSK; In-silico and biochemical measurements: PA & SSA; Data analysis: SSA & PA; Paper writing and Figure preparation: PA, & SSA; Statistical analysis: PA & SSA; Revision, English editing & approval of the final paper: JI, MSK & SSA.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ahmad, P., Alvi, S.S., Iqbal, J. et al. Identification and evaluation of natural organosulfur compounds as potential dual inhibitors of α-amylase and α-glucosidase activity: an in-silico and in-vitro approach. Med Chem Res 30, 2184–2202 (2021). https://doi.org/10.1007/s00044-021-02799-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-021-02799-2